Epigenetic Changes in Asthma: Role of DNA CpG Methylation
- PMID: 31905427
- PMCID: PMC6953489
- DOI: 10.4046/trd.2018.0088
Epigenetic Changes in Asthma: Role of DNA CpG Methylation
Abstract
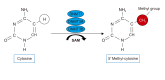
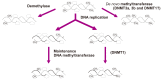
For the past three decades, more than a thousand of genetic studies have been performed to find out the genetic variants responsible for the risk of asthma. Until now, all of the discovered single nucleotide polymorphisms have explained genetic effects less than initially expected. Thus, clarification of environmental factors has been brought up to overcome the 'missing' heritability. The most exciting solution is epigenesis because it intervenes at the junction between the genome and the environment. Epigenesis is an alteration of genetic expression without changes of DNA sequence caused by environmental factors such as nutrients, allergens, cigarette smoke, air pollutants, use of drugs and infectious agents during pre- and post-natal periods and even in adulthood. Three major forms of epigenesis are composed of DNA methylation, histone modifications, and specific microRNA. Recently, several studies have been published on epigenesis in asthma and allergy as a powerful tool for research of genetic heritability in asthma albeit epigenetic changes are at the starting point to obtain the data on specific phenotypes of asthma. In this presentation, we mainly review the potential role of DNA CpG methylation in the risk of asthma and its sub-phenotypes including nonsteroidal anti-inflammatory exacerbated respiratory diseases.
Keywords: Aspirin; Asthma; Environment; Epigenesis; Gene.
Copyright©2020. The Korean Academy of Tuberculosis and Respiratory Diseases.
Conflict of interest statement
No potential conflict of interest relevant to this article was reported.
Figures

References
-
- Edfors-Lubs ML. Allergy in 7000 twin pairs. Acta Allergol. 1971;26:249–285. - PubMed
-
- Moffatt MF, Kabesch M, Liang L, Dixon AL, Strachan D, Heath S, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448:470–473. - PubMed
-
- Vercelli D. Learning from discrepancies: CD14 polymorphisms, atopy and the endotoxin switch. Clin Exp Allergy. 2003;33:153–155. - PubMed
-
- Bouzigon E, Corda E, Aschard H, Dizier MH, Boland A, Bousquet J, et al. Effect of 17q21 variants and smoking exposure in early-onset asthma. N Engl J Med. 2008;359:1985–1994. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

