Histone 2A Family Member J Drives Mesenchymal Transition and Temozolomide Resistance in Glioblastoma Multiforme
- PMID: 31906036
- PMCID: PMC7016639
- DOI: 10.3390/cancers12010098
Histone 2A Family Member J Drives Mesenchymal Transition and Temozolomide Resistance in Glioblastoma Multiforme
Erratum in
-
Erratum: Lee, H.H., et al. Histone 2A Family Member J Drives Mesenchymal Transition and Temozolomide Resistance in Glioblastoma Multiforme. Cancers 2020, 12, 98.Cancers (Basel). 2020 Sep 16;12(9):2649. doi: 10.3390/cancers12092649. Cancers (Basel). 2020. PMID: 32948061 Free PMC article.
Abstract
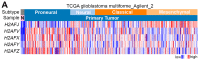
Glioblastoma multiforme (GBM) is the most aggressive brain tumor and has a poor prognosis and is poorly sensitive to radiotherapy or temozolomide (TMZ) chemotherapy. Therefore, identifying new biomarkers to predict therapeutic responses of GBM is urgently needed. By using The Cancer Genome Atlas (TCGA) database, we found that the upregulation of histone 2A family member J (H2AFJ), but not other H2AFs, is extensively detected in the therapeutic-insensitive mesenchymal, IDH wildtype, MGMT unmethylated, or non-G-CIMP GBM and is associated with poor TMZ responsiveness independent of radiation. Similar views were also found in GBM cell lines. Whereas H2AFJ knockdown diminished TMZ resistance, H2AFJ overexpression promoted TMZ resistance in a panel of GBM cell lines. Gene set enrichment analysis (GSEA) revealed that H2AFJ upregulation accompanied by the activation of TNF-α/NF-κB and IL-6/STAT3-related pathways is highly predicted. Luciferase-based promoter activity assay further validated that the activities of NF-κB and STAT3 are causally affected by H2AFJ expression in GBM cells. Moreover, we found that therapeutic targeting HADC3 by tacedinaline or NF-κB by ML029 is likely able to overcome the TMZ resistance in GBM cells with H2AFJ upregulation. Significantly, the GBM cohorts harboring a high-level H2AFJ transcript combined with high-level expression of TNF-α/NF-κB geneset, IL-6/STAT3 geneset or HADC3 were associated with a shorter time to tumor repopulation after initial treatment with TMZ. These findings not only provide H2AFJ as a biomarker to predict TMZ therapeutic effectiveness but also suggest a new strategy to combat TMZ-insensitive GBM by targeting the interaction network constructed by TNF-α/NF-κB, IL-6/STAT3, HDAC3, and H2AFJ.
Keywords: GBM; H2AFJ; MGMT; Temozolomide; precision medicine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures









References
-
- Louis D.N., Perry A., Reifenberger G., von D.A., Figarella-Branger D., Cavenee W.K., Ohgaki H., Wiestler O.D., Kleihues P., Ellison D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. - DOI - PubMed
-
- Chandana S.R., Movva S., Arora M., Singh T. Primary brain tumors in adults. Am. Fam. Physician. 2008;77:1423–1430. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

