A Single Cell but Many Different Transcripts: A Journey into the World of Long Non-Coding RNAs
- PMID: 31906285
- PMCID: PMC6982300
- DOI: 10.3390/ijms21010302
A Single Cell but Many Different Transcripts: A Journey into the World of Long Non-Coding RNAs
Abstract
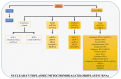
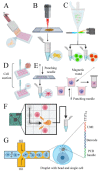
In late 2012 it was evidenced that most of the human genome is transcribed but only a small percentage of the transcripts are translated. This observation supported the importance of non-coding RNAs and it was confirmed in several organisms. The most abundant non-translated transcripts are long non-coding RNAs (lncRNAs). In contrast to protein-coding RNAs, they show a more cell-specific expression. To understand the function of lncRNAs, it is fundamental to investigate in which cells they are preferentially expressed and to detect their subcellular localization. Recent improvements of techniques that localize single RNA molecules in tissues like single-cell RNA sequencing and fluorescence amplification methods have given a considerable boost in the knowledge of the lncRNA functions. In recent years, single-cell transcription variability was associated with non-coding RNA expression, revealing this class of RNAs as important transcripts in the cell lineage specification. The purpose of this review is to collect updated information about lncRNA classification and new findings on their function derived from single-cell analysis. We also retained useful for all researchers to describe the methods available for single-cell analysis and the databases collecting single-cell and lncRNA data. Tables are included to schematize, describe, and compare exposed concepts.
Keywords: lncRNA database; lncRNAs; long non-coding RNAs; non-coding RNAs; single-cell; single-cell database; single-cell expression; single-cell sequencing.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

