Analysis of Small RNAs of Barley Genotypes Associated with Resistance to Barley Yellow Dwarf Virus
- PMID: 31906504
- PMCID: PMC7020447
- DOI: 10.3390/plants9010060
Analysis of Small RNAs of Barley Genotypes Associated with Resistance to Barley Yellow Dwarf Virus
Abstract
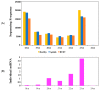
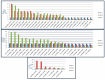
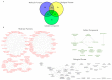
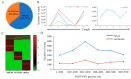
Barley yellow dwarf virus (BYDV) causes an often-devastating disease of cereals that is most effectively controlled by using plant genotypes that are resistant or tolerant to the virus. New barley lines Vir8:3 and Vir13:8, with pyramided resistance genes against different pathogens and resistance gene Ryd2 against BYDV, are currently being tested. Because microRNAs (miRNAs) are associated with antiviral plant defense, here we compared the miRNA profiles in these lines and in cultivar Wysor (carrying one resistance gene, Ryd2), with and without BYDV infection and after feeding by virus-free aphids, to determine whether the miRNA profile in the resistant variety bear similarities with the newly developed lines. The BYDV titer for each group was also determined and compared to the titer in sensitive cultivar Graciosa. Among 746 miRNAs identified in barley, 66 were known miRNAs, and 680 were novel. The expression of 73 miRNAs differed significantly after BYDV infection, including the strong, specific upregulation of novel miRNA10778 that was conserved across all the barley genotypes. This miRNA belongs to the H box and ACA box (H/ACA) snoR14 family of RNAs (Rf01280) and is associated with pseudourydilation. The expression of 48 miRNAs also differed depending on the barley genotype. The profile of miRNAs expressed in Vir8:3 and Vir13:8 in response to BYDV was similar and differed from that of Wysor. Insights into the expression patterns of miRNAs in response to BYDV in barley provided here will benefit further studies toward understanding the resistance mechanisms and developing novel strategies against virus infections.
Keywords: BYDV; NGS; Ryd2; miRNA; sRNA; sequencing.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Identifying Putative Resistance Genes for Barley Yellow Dwarf Virus-PAV in Wheat and Barley.Viruses. 2023 Mar 9;15(3):716. doi: 10.3390/v15030716. Viruses. 2023. PMID: 36992425 Free PMC article. Review.
-
Pyramiding of Ryd2 and Ryd3 conferring tolerance to a German isolate of Barley yellow dwarf virus-PAV (BYDV-PAV-ASL-1) leads to quantitative resistance against this isolate.Theor Appl Genet. 2011 Jun;123(1):69-76. doi: 10.1007/s00122-011-1567-y. Epub 2011 Mar 18. Theor Appl Genet. 2011. PMID: 21416402
-
Barley yellow dwarf virus Infection Leads to Higher Chemical Defense Signals and Lower Electrophysiological Reactions in Susceptible Compared to Tolerant Barley Genotypes.Front Plant Sci. 2018 Mar 6;9:145. doi: 10.3389/fpls.2018.00145. eCollection 2018. Front Plant Sci. 2018. PMID: 29563918 Free PMC article.
-
Agronomical, biochemical and histological response of resistant and susceptible wheat and barley under BYDV stress.PeerJ. 2018 May 28;6:e4833. doi: 10.7717/peerj.4833. eCollection 2018. PeerJ. 2018. PMID: 29868264 Free PMC article.
-
Molecular markers in breeding for virus resistance in barley.J Appl Genet. 2004;45(2):145-59. J Appl Genet. 2004. PMID: 15131346 Review.
Cited by
-
Control of Plant Viral Diseases by CRISPR/Cas9: Resistance Mechanisms, Strategies and Challenges in Food Crops.Plants (Basel). 2021 Jun 22;10(7):1264. doi: 10.3390/plants10071264. Plants (Basel). 2021. PMID: 34206201 Free PMC article. Review.
-
Identifying Putative Resistance Genes for Barley Yellow Dwarf Virus-PAV in Wheat and Barley.Viruses. 2023 Mar 9;15(3):716. doi: 10.3390/v15030716. Viruses. 2023. PMID: 36992425 Free PMC article. Review.
References
-
- Adams M.J., Lefkowitz E.J., King A.M.Q., Harrach B., Harrison R.L., Knowles N.J., Kropinski A.M., Krupovic M., Kuhn J.H., Mushegian A.R. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses. Arch. Virol. 2016;161:2921–2949. doi: 10.1007/s00705-016-2977-6. - DOI - PMC - PubMed
-
- D’Arcy C.J., Burnett P.A. Barley Yellow Dwarf: 40 Years of Progress. APS Press; St Paul, MN, USA: 1995. 374p
-
- Pike K.S. A review of barley yellow dwarf virus grain losses. In: Burnett P.A., editor. World Perspectives on Barley Yellow Dwarf Virus. International Maize and Wheat Improvement Center; Mexico City, Mexico: 1990. pp. 356–359.
-
- Henry M., Posadas G., Segura J., Rajaram S. Evaluating resistance to BYDV-PAV, BYDV-MAV, and CYDV-RPV in Thinopyrum intermedium-derived wheat lines; Proceedings of the International Symposium; Texcoco, Mexico. 1–5 September 2002; pp. 64–66.
Grants and funding
LinkOut - more resources
Full Text Sources

