Differential Subcellular Distribution and Translocation of Seven 14-3-3 Isoforms in Response to EGF and During the Cell Cycle
- PMID: 31906564
- PMCID: PMC6981507
- DOI: 10.3390/ijms21010318
Differential Subcellular Distribution and Translocation of Seven 14-3-3 Isoforms in Response to EGF and During the Cell Cycle
Abstract
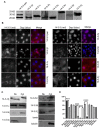
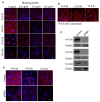
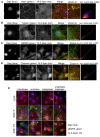
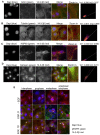
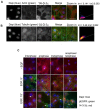
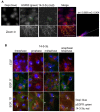
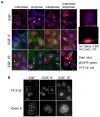
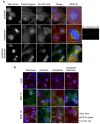
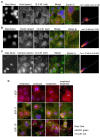
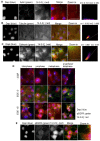
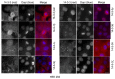
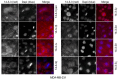
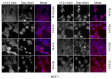
Multiple isoforms of 14-3-3 proteins exist in different organisms. In mammalian cells, 14-3-3 protein has seven isoforms (α/β, ε, η, γ, σ, θ/τ, and δ/ζ), with α and δ representing the phosphorylated versions of β and ζ, respectively. While the existence of multiple isoforms may represent one more level of regulation in 14-3-3 signaling, our knowledge regarding the isoform-specific functions of 14-3-3 proteins is very limited. Determination of the subcellular localization of the different 14-3-3 isoforms could give us important clues of their specific functions. In this study, by using indirect immunofluorescence, subcellular fractionation, and immunoblotting, we studied the subcellular localization of the total 14-3-3 protein and each of the seven 14-3-3 isoforms; their redistribution throughout the cell cycle; and their translocation in response to EGF in Cos-7 cells. We showed that 14-3-3 proteins are broadly distributed throughout the cell and associated with many subcellular structures/organelles, including the plasma membrane (PM), mitochondria, ER, nucleus, microtubules, and actin fibers. This broad distribution underlines the multiple functions identified for 14-3-3 proteins. The different isoforms of 14-3-3 proteins have distinctive subcellular localizations, which suggest their distinctive cellular functions. Most notably, 14-3-3ƞ is almost exclusively localized to the mitochondria, 14-3-3γ is only localized to the nucleus, and 14-3-3σ strongly and specifically associated with the centrosome during mitosis. We also examined the subcellular localization of the seven 14-3-3 isoforms in other cells, including HEK-293, MDA-MB-231, and MCF-7 cells, which largely confirmed our findings with Cos-7 cells.
Keywords: 14-3-3 proteins; actin fibers; and microtubules; centrosome; isoforms; mitochondria; nucleus; subcellular localization.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Rac1 S71 Mediates the Interaction between Rac1 and 14-3-3 Proteins.Cells. 2019 Aug 30;8(9):1006. doi: 10.3390/cells8091006. Cells. 2019. PMID: 31480268 Free PMC article.
-
Isoform-specific differences in rapid nucleocytoplasmic shuttling cause distinct subcellular distributions of 14-3-3 sigma and 14-3-3 zeta.J Cell Sci. 2004 Mar 15;117(Pt 8):1411-20. doi: 10.1242/jcs.00990. Epub 2004 Mar 2. J Cell Sci. 2004. PMID: 14996909
-
Cell Cycle-dependent Changes in Localization and Phosphorylation of the Plasma Membrane Kv2.1 K+ Channel Impact Endoplasmic Reticulum Membrane Contact Sites in COS-1 Cells.J Biol Chem. 2015 Dec 4;290(49):29189-201. doi: 10.1074/jbc.M115.690198. Epub 2015 Oct 6. J Biol Chem. 2015. PMID: 26442584 Free PMC article.
-
The functional significance of 14-3-3 proteins in cancer: focus on lung cancer.Horm Mol Biol Clin Investig. 2017 Aug 5;32(3):/j/hmbci.2017.32.issue-3/hmbci-2017-0032/hmbci-2017-0032.xml. doi: 10.1515/hmbci-2017-0032. Horm Mol Biol Clin Investig. 2017. PMID: 28779564 Review.
-
Structural determinants of 14-3-3 binding specificities and regulation of subcellular localization of 14-3-3-ligand complexes: a comparison of the X-ray crystal structures of all human 14-3-3 isoforms.Semin Cancer Biol. 2006 Jun;16(3):173-82. doi: 10.1016/j.semcancer.2006.03.007. Epub 2006 Apr 1. Semin Cancer Biol. 2006. PMID: 16678437 Review.
Cited by
-
Probing the 14-3-3 Isoform-Specificity Profile of Protein-Protein Interactions Stabilized by Fusicoccin A.ACS Omega. 2020 Sep 24;5(39):25029-25035. doi: 10.1021/acsomega.0c01454. eCollection 2020 Oct 6. ACS Omega. 2020. PMID: 33043180 Free PMC article.
-
Clinical significance of serum levels of 14-3-3β protein in patients with stable chronic obstructive pulmonary disease.Sci Rep. 2023 Mar 24;13(1):4861. doi: 10.1038/s41598-023-32096-4. Sci Rep. 2023. PMID: 36964173 Free PMC article.
-
Structural insights into regulation of the PEAK3 pseudokinase scaffold by 14-3-3.Nat Commun. 2023 Jun 19;14(1):3543. doi: 10.1038/s41467-023-38864-0. Nat Commun. 2023. PMID: 37336883 Free PMC article.
-
Evidence of 14-3-3 proteins contributing to kinetochore integrity and chromosome congression during mitosis.J Cell Sci. 2024 Aug 1;137(15):jcs261928. doi: 10.1242/jcs.261928. Epub 2024 Aug 9. J Cell Sci. 2024. PMID: 38988319 Free PMC article.
-
Regulation of H9C2 cell hypertrophy by 14-3-3η via inhibiting glycolysis.PLoS One. 2024 Jul 22;19(7):e0307696. doi: 10.1371/journal.pone.0307696. eCollection 2024. PLoS One. 2024. PMID: 39038022 Free PMC article.
References
-
- Thomas D., Guthridge M., Woodcock J., Lopez A. 14-3-3 protein Signal. in development and growth factor responses. Curr. Top. Dev. Biol. 2005;67:285–303. - PubMed
-
- Ichimura T., Isobe T., Okuyama T., Takahashi N., Araki K., Kuwano R., Takahashi Y. Molecular cloning of cDNA coding for brain-specific 14-3-3 protein, a protein kinase-dependent activator of tyrosine and tryptophan hydroxylases. Proc. Natl. Acad. Sci. USA. 1988;85:7084–7088. doi: 10.1073/pnas.85.19.7084. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

