Loss of the SWI/SNF-ATPase subunit members SMARCF1 (ARID1A), SMARCA2 (BRM), SMARCA4 (BRG1) and SMARCB1 (INI1) in oesophageal adenocarcinoma
- PMID: 31906887
- PMCID: PMC6945480
- DOI: 10.1186/s12885-019-6425-3
Loss of the SWI/SNF-ATPase subunit members SMARCF1 (ARID1A), SMARCA2 (BRM), SMARCA4 (BRG1) and SMARCB1 (INI1) in oesophageal adenocarcinoma
Abstract
Background: The SWI/SNF complex is an important chromatin remodeler, commonly dysregulated in cancer, with an estimated mutation frequency of 20%. ARID1A is the most frequently mutated subunit gene. Almost nothing is known about the other familiar members of the SWI/SNF complexes, SMARCA2 (BRM), SMARCA4 (BRG1) and SMARCB1 (INI1), in oesophageal adenocarcinoma (EAC).
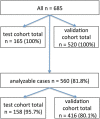
Methods: We analysed a large cohort of 685 patients with EAC. We used four different antibodies to detect a loss-of-protein of ARID1A BRM, BRG1 and INI1 by immunohistochemistry and correlated these findings with molecular and clinical data.
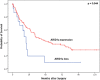
Results: Loss of ARID1A, BRG1, BRM and INI1 was observed in 10.4, 3.4, 9.9 and 2% of EAC. We found a co-existing protein loss of ARID1A and BRM in 9.9% and of ARID1A and BRG1 in 2.2%. Patients with loss of ARID1A and TP53 wildtype EACs showed a shortened overall survival compared with AIRDA1A-positive tumours [median overall survival was 60.1 months (95%CI 1.2-139.9 months)] in patients with ARIDA-1A expression and 26.2 months (95%CI 3.7-19.1 months) in cases of ARIDA-1A loss (p = 0.044). Tumours with loss or expression of ARID1A and TP53 loss were not associated with a difference in survival. Only one tumour revealed high microsatellite instability (MSI-H) with concomitant ARID1A loss. All other ARID1A loss-EACs were microsatellite-stable (MSS). No predictive relevance was seen for SWI/SNF-complex alterations and simultaneous amplification of different genes (PIK3CA, KRAS, c-MYC, MET, GATA6, ERBB2).
Conclusion: Our work describes, for the first time, loss of one of the SWI/SNF ATPase subunit proteins in a large number of adenocarcinomas of the oesophagus. Several papers discuss possible therapeutic interventions for tumours showing a loss of function of the SWI/SNF complex, such as PARP inhibitors or PI3K and AKT inhibitors. Future studies will be needed to show whether SWI/SNF complex-deficient EACs may benefit from personalized therapy.
Keywords: Heterogeneity; Loss-of-function; Oesophageal adenocarcinoma; SWI/SNF-complex; TP53 loss.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures




Similar articles
-
Loss of SWI/SNF complex expression (SMARCA4, SMARCA2, SMARCB1, ARID1A) is associated with dMMR in primary adenocarcinoma of jejunum and ileum: A clinicopathological and molecular analysis based on the Chinese population.Pathol Res Pract. 2025 May;269:155891. doi: 10.1016/j.prp.2025.155891. Epub 2025 Mar 11. Pathol Res Pract. 2025. PMID: 40101550
-
Dual loss of the SWI/SNF complex ATPases SMARCA4/BRG1 and SMARCA2/BRM is highly sensitive and specific for small cell carcinoma of the ovary, hypercalcaemic type.J Pathol. 2016 Feb;238(3):389-400. doi: 10.1002/path.4633. Epub 2015 Dec 21. J Pathol. 2016. PMID: 26356327 Free PMC article.
-
Switch/sucrose-non-fermentable (SWI/SNF) complex (SMARCA4, SMARCA2, INI1/SMARCB1)-deficient colorectal carcinomas are strongly associated with microsatellite instability: an incidence study in 4508 colorectal carcinomas.Histopathology. 2022 May;80(6):906-921. doi: 10.1111/his.14612. Epub 2022 Feb 24. Histopathology. 2022. PMID: 34951482
-
Chromatin remodeling and cancer: the critical influence of the SWI/SNF complex.Epigenetics Chromatin. 2025 Apr 23;18(1):22. doi: 10.1186/s13072-025-00590-w. Epigenetics Chromatin. 2025. PMID: 40269969 Free PMC article. Review.
-
[Clinicopathological features of gastric alpha-fetoprotein-producing adenocarcinoma with SWI/SNF complex deletion].Zhonghua Bing Li Xue Za Zhi. 2024 Jan 8;53(1):52-57. doi: 10.3760/cma.j.cn112151-20231023-00284. Zhonghua Bing Li Xue Za Zhi. 2024. PMID: 38178747 Review. Chinese.
Cited by
-
Combined score of Y chromosome loss and T-cell infiltration improves UICC based stratification of esophageal adenocarcinoma.Front Oncol. 2023 Nov 17;13:1249172. doi: 10.3389/fonc.2023.1249172. eCollection 2023. Front Oncol. 2023. PMID: 38045001 Free PMC article.
-
SMARCA4 Mutations in Gastroesophageal Adenocarcinoma: An Observational Study via a Next-Generation Sequencing Panel.Cancers (Basel). 2024 Mar 27;16(7):1300. doi: 10.3390/cancers16071300. Cancers (Basel). 2024. PMID: 38610978 Free PMC article.
-
SWI/SNF Complex-Deficient Undifferentiated Carcinoma of the Pancreas: Clinicopathologic and Genomic Analysis.Mod Pathol. 2024 Nov;37(11):100585. doi: 10.1016/j.modpat.2024.100585. Epub 2024 Jul 31. Mod Pathol. 2024. PMID: 39094734 Clinical Trial.
-
Toward Targeted Therapies in Oesophageal Cancers: An Overview.Cancers (Basel). 2022 Mar 16;14(6):1522. doi: 10.3390/cancers14061522. Cancers (Basel). 2022. PMID: 35326673 Free PMC article. Review.
-
Metastatic SMARCA4-deficient undifferentiated tumor in the small mesentery: case report.World J Surg Oncol. 2024 Dec 20;22(1):337. doi: 10.1186/s12957-024-03619-8. World J Surg Oncol. 2024. PMID: 39707357 Free PMC article.
References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

