Evidence for phylogenetically and catabolically diverse active diazotrophs in deep-sea sediment
- PMID: 31907368
- PMCID: PMC7082343
- DOI: 10.1038/s41396-019-0584-8
Evidence for phylogenetically and catabolically diverse active diazotrophs in deep-sea sediment
Abstract
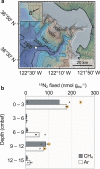
Diazotrophic microorganisms regulate marine productivity by alleviating nitrogen limitation. However, we know little about the identity and activity of diazotrophs in deep-sea sediments, a habitat covering nearly two-thirds of the planet. Here, we identify candidate diazotrophs from Pacific Ocean sediments collected at 2893 m water depth using 15N-DNA stable isotope probing and a novel pipeline for nifH sequence analysis. Together, these approaches detect an unexpectedly diverse assemblage of active diazotrophs, including members of the Acidobacteria, Firmicutes, Nitrospirae, Gammaproteobacteria, and Deltaproteobacteria. Deltaproteobacteria, predominately members of the Desulfobacterales and Desulfuromonadales, are the most abundant diazotrophs detected, and display the most microdiversity of associated nifH sequences. Some of the detected lineages, including those within the Acidobacteria, have not previously been shown to fix nitrogen. The diazotrophs appear catabolically diverse, with the potential for using oxygen, nitrogen, iron, sulfur, and carbon as terminal electron acceptors. Therefore, benthic diazotrophy may persist throughout a range of geochemical conditions and provide a stable source of fixed nitrogen over geologic timescales. Our results suggest that nitrogen-fixing communities in deep-sea sediments are phylogenetically and catabolically diverse, and open a new line of inquiry into the ecology and biogeochemical impacts of deep-sea microorganisms.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures




References
-
- Galloway JN, Dentener FJ, Capone DG, Boyer EW, Howarth RW, Seitzinger SP, et al. Nitrogen cycles: past, present, and future. Biogeochemistry. 2004;70:153–226. doi: 10.1007/s10533-004-0370-0. - DOI
-
- Howarth RW. Nutrient limitation of net primary production in marine ecosystems. Annu Rev Ecol Syst. 1988;19:89–110. doi: 10.1146/annurev.es.19.110188.000513. - DOI

