Genomic evidence supports a clonal diaspora model for metastases of esophageal adenocarcinoma
- PMID: 31907488
- PMCID: PMC7100916
- DOI: 10.1038/s41588-019-0551-3
Genomic evidence supports a clonal diaspora model for metastases of esophageal adenocarcinoma
Abstract
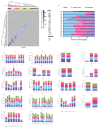
The poor outcomes in esophageal adenocarcinoma (EAC) prompted us to interrogate the pattern and timing of metastatic spread. Whole-genome sequencing and phylogenetic analysis of 388 samples across 18 individuals with EAC showed, in 90% of patients, that multiple subclones from the primary tumor spread very rapidly from the primary site to form multiple metastases, including lymph nodes and distant tissues-a mode of dissemination that we term 'clonal diaspora'. Metastatic subclones at autopsy were present in tissue and blood samples from earlier time points. These findings have implications for our understanding and clinical evaluation of EAC.
Conflict of interest statement
The authors declare no competing interests.
Figures




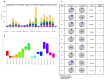
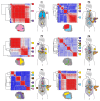
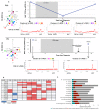
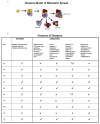
References
Publication types
MeSH terms
Supplementary concepts
Grants and funding
- 22131/CRUK_/Cancer Research UK/United Kingdom
- P30 CA023100/CA/NCI NIH HHS/United States
- MR/K00316X/1/MRC_/Medical Research Council/United Kingdom
- A15874/CRUK_/Cancer Research UK/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MC_UU_12022/2/MRC_/Medical Research Council/United Kingdom
- 15874/CRUK_/Cancer Research UK/United Kingdom
- MC_EX_UU_MR/K00316X/1/MRC_/Medical Research Council/United Kingdom
- 21777/CRUK_/Cancer Research UK/United Kingdom
- A12770/CRUK_/Cancer Research UK/United Kingdom
- 22720/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

