Control of Rab7a activity and localization through endosomal type Igamma PIP 5-kinase is required for endosome maturation and lysosome function
- PMID: 31908013
- PMCID: PMC7018547
- DOI: 10.1096/fj.201901830R
Control of Rab7a activity and localization through endosomal type Igamma PIP 5-kinase is required for endosome maturation and lysosome function
Abstract
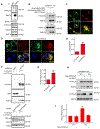
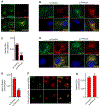
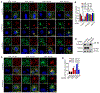
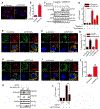
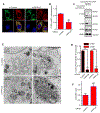
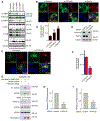
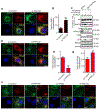
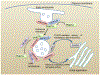
The small GTPase Ras-related protein Rab-7a (Rab7a) serves as a key organizer of the endosomal-lysosomal system. However, molecular mechanisms controlling Rab7a activation levels and subcellular translocation are still poorly defined. Here, we demonstrate that type Igamma phosphatidylinositol phosphate 5-kinase i5 (PIPKIγi5), an endosome-localized enzyme that produces phosphatidylinositol 4,5-bisphosphate, directly interacts with Rab7a and plays critical roles in the control of the endosomal-lysosomal system. The loss of PIPKIγi5 blocks Rab7a recruitment to early endosomes, which prevents the maturation of early to late endosomes. PIPKIγi5 loss disturbs retromer complex connection with Rab7a, which blocks the retrograde sorting of Cation-independent Mannose 6-Phosphate Receptor from late endosomes. This leads to the decreased sorting of hydrolases to lysosomes and reduces the autophagic degradation. By modulating the retromer-Rab7a connection, PIPKIγi5 is also required for the recruitment of the GTPase-activating protein TBC1 domain family member 5 to late endosomes, which controls the conversion of Rab7a from the active state to the inactive state. Thus, PIPKIγi5 is critical for the modulation of Rab7a activity, localization, and function in the endosomal-lysosomal system.
Keywords: PIPKIγi5; Rab7a; autophagy; endosome maturation; lysosome.
© 2019 Federation of American Societies for Experimental Biology.
Figures
Similar articles
-
Endosomal type Iγ PIP 5-kinase controls EGF receptor lysosomal sorting.Dev Cell. 2013 Apr 29;25(2):144-55. doi: 10.1016/j.devcel.2013.03.010. Epub 2013 Apr 18. Dev Cell. 2013. PMID: 23602387 Free PMC article.
-
Inhibition of TBC1D5 activates Rab7a and can enhance the function of the retromer cargo-selective complex.J Cell Sci. 2018 Jun 21;131(12):jcs217398. doi: 10.1242/jcs.217398. J Cell Sci. 2018. PMID: 29777037 Free PMC article.
-
Involvement of CASP9 (caspase 9) in IGF2R/CI-MPR endosomal transport.Autophagy. 2021 Jun;17(6):1393-1409. doi: 10.1080/15548627.2020.1761742. Epub 2020 May 25. Autophagy. 2021. PMID: 32397873 Free PMC article.
-
Rab7a and Mitophagosome Formation.Cells. 2019 Mar 8;8(3):224. doi: 10.3390/cells8030224. Cells. 2019. PMID: 30857122 Free PMC article. Review.
-
Rab GTPases regulating receptor trafficking at the late endosome-lysosome membranes.Cell Biochem Funct. 2012 Aug;30(6):515-23. doi: 10.1002/cbf.2827. Epub 2012 Apr 3. Cell Biochem Funct. 2012. PMID: 22473705 Review.
Cited by
-
Phosphatidylinositol 4,5-bisphosphate in the Control of Membrane Trafficking.Int J Biol Sci. 2020 Aug 25;16(15):2761-2774. doi: 10.7150/ijbs.49665. eCollection 2020. Int J Biol Sci. 2020. PMID: 33061794 Free PMC article. Review.
-
Ca2+ and Annexins - Emerging Players for Sensing and Transferring Cholesterol and Phosphoinositides via Membrane Contact Sites.Adv Exp Med Biol. 2023;1422:393-438. doi: 10.1007/978-3-031-21547-6_15. Adv Exp Med Biol. 2023. PMID: 36988890
-
PIP5K1C phosphoinositide kinase deficiency distinguishes PIKFYVE-dependent cancer cells from non-malignant cells.Autophagy. 2023 Sep;19(9):2464-2484. doi: 10.1080/15548627.2023.2182594. Epub 2023 Mar 22. Autophagy. 2023. PMID: 36803256 Free PMC article.
-
Integrative clinical and molecular characterization of translocation renal cell carcinoma.Cell Rep. 2022 Jan 4;38(1):110190. doi: 10.1016/j.celrep.2021.110190. Cell Rep. 2022. PMID: 34986355 Free PMC article.
-
The secreted protein kinase CstK from Coxiella burnetii influences vacuole development and interacts with the GTPase-activating host protein TBC1D5.J Biol Chem. 2020 May 22;295(21):7391-7403. doi: 10.1074/jbc.RA119.010112. Epub 2020 Apr 17. J Biol Chem. 2020. PMID: 32303638 Free PMC article.
References
-
- Cullen PJ, and Steinberg F (2018) To degrade or not to degrade: mechanisms and significance of endocytic recycling. Nature reviews. Molecular cell biology 19, 679–696 - PubMed
-
- McNally KE, and Cullen PJ (2018) Endosomal Retrieval of Cargo: Retromer Is Not Alone. Trends in cell biology 28, 807–822 - PubMed
-
- Podinovskaia M, and Spang A (2018) The Endosomal Network: Mediators and Regulators of Endosome Maturation. Progress in molecular and subcellular biology 57, 1–38 - PubMed
-
- Braulke T, and Bonifacino JS (2009) Sorting of lysosomal proteins. Biochimica et biophysica acta 1793, 605–614 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

