PCAF-mediated acetylation of ISX recruits BRD4 to promote epithelial-mesenchymal transition
- PMID: 31908141
- PMCID: PMC7001155
- DOI: 10.15252/embr.201948795
PCAF-mediated acetylation of ISX recruits BRD4 to promote epithelial-mesenchymal transition
Abstract
Epigenetic regulation is important for cancer progression; however, the underlying mechanisms, particularly those involving protein acetylation, remain to be fully understood. Here, we show that p300/CBP-associated factor (PCAF)-dependent acetylation of the transcription factor intestine-specific homeobox (ISX) regulates epithelial-mesenchymal transition (EMT) and promotes cancer metastasis. Mechanistically, PCAF acetylation of ISX at lysine 69 promotes the interaction with acetylated bromodomain-containing protein 4 (BRD4) at lysine 332 in tumor cells, and the translocation of the resulting complex into the nucleus. There, it binds to promoters of EMT genes, where acetylation of histone 3 at lysines 9, 14, and 18 initiates chromatin remodeling and subsequent transcriptional activation. Ectopic ISX expression enhances EMT marker expression, including TWIST1, Snail1, and VEGF, induces cancer metastasis, but suppresses E-cadherin expression. In lung cancer, ectopic expression of PCAF-ISX-BRD4 axis components correlates with clinical metastatic features and poor prognosis. These results suggest that the PCAF-ISX-BRD4 axis mediates EMT signaling and regulates tumor initiation and metastasis.
Keywords: EMT; ISX; BRD4; TWIST1.
© 2020 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

- A
The expression levels of ISX, TWIST1, Snail1, fibronectin (FN1), VEGF, and E‐cadherin (CDH1) mRNA were examined in a ISX‐GFP inducible Tet‐ON transformants at 8 h after addition of DOX. Data are presented as mean ± SD in graph (P < 0.001 compared with time point 0 h; Student's t‐test) of three independent experiments, each performed in triplicate.
- B
Western blotting analysis of the protein levels of ISX, HIF1α, TWIST1, Snail1, Slug, ZEB1, Bmi1, E‐cadherin (E‐Cad.), fibronectin, N‐cadherin (N‐Cad.), vimentin, and VEGF in A549 and H1299 cells with DOX‐inducible ISX expression system 8 h after DOX induction.
- C, D
ISX transcriptionally activates luciferase activity driven by TWIST1 (C) and Snail1 (D) promoter regions in A549 cells. Data are presented as mean ± SD in bar graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- E
ChIP analysis of ISX binding to the TWIST1 promoter region in A549 and H1299 cells (10% input of each group was pull down and applied to qPCR). Data are presented as mean ± SD in bar graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- F
ISX transactivation activity analyzed by luciferase activity driven by the TWIST1 promoter. Data are presented as mean ± SD in graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- G
ChIP analysis of ISX binding to the endogenous promoters of TWIST1 (−180/+35) and Snail1(−160/+40) in A549 and cells (10% input of each group was pull down and applied to qPCR). Data are presented as mean ± SD in bar graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- H
Effect of forced expression and knockdown of ISX on cell migration (wound healing) measured in A549 cells. ISXi, ISX‐specific shRNAi (described in Materials and Methods). Data are presented as mean ± SD in graph (**P < 0.01, ***P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate. Scale bar, 100 μm.
- I
Effect of forced expression and knockdown of ISX on cell invasion (Transwell) activity measured in A549 cells. Data are presented as mean ± SD in bar graph (***P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate. Scale bar, 100 μm.

- A
ChIP analysis of ISX binding to the promoters of Snail1 in A549 and H1299 cells. Data are presented as mean ± SD in bar graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- B
ISX transactivation activity analyzed by luciferase activity driven by the Snail1 promoter. Data are presented as mean ± SD in graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- C
ISX‐associated proteins (PCAF, BRD4, CBP, CREB, and histone H3) determined by Western blotting in immunoprecipitation of H1299 cells.
- D, E
Endogenous mRNA and protein expression of ISX, PCAF, and BRD4 was determined in various lung cancer cell lines. Data are presented as mean ± SD in graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- F
Confocal immunofluorescence detection of ISX (green), BRD4 (red), and PCAF (pink) in A549 cells. Cell nuclei were visualized by DAPI (blue). Blank arrow, co‐localization.
- G
Confocal immunofluorescence detection of ISX (green) and BRD4 (red) localization in A549 cells treated with Garcinol. Cell nuclei were visualized by DAPI (blue). Yellow arrows indicate co‐localization in nuclei, and white arrows indicate co‐localization in the cytosol.
- H
A quantification of ISX in cytosol and nuclei. Data are presented as mean ± SD in bar graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- I
PCAF expression in A549 cells was knocked down with 7 sequence‐specific shRNAi constructs.

- A
BRD4 peptide (291–314 a.a) obtained from anti‐ISX immunoprecipitates of A549 cell lysates identified through liquid chromatography–tandem mass spectrometry (LC‐tandem‐MS).
- B
ISX association proteins (PCAF, BRD4, CBP, CREB, RNA Pol II, and Histone H3) determined by Western blot in immunoprecipitation of A549 cells.
- C
ISX association proteins (PCAF, BRD4, CBP, CREB, and RNA Pol II) determined by Western blot in anti‐ISX immunoprecipitates of tumor tissues from patients with lung cancer.
- D
Proximity ligation assay (PLA) of ISX and BRD4(PCAF) interaction in A549 cells. Red foci indicate close proximity of the two proteins. NC: negative control. Blank arrows indicate interactions of ISX with BRD4 (PCAF) in the cells.
- E
Western blotting analysis of the protein levels of ISX, PCAF, and EMT markers in DOX‐inducible ISX expression system A549 cells treated with TH1834, Garcinol, C646, and MB‐3 for 8 h after DOX induction.
- F
Proximity ligation assay of ISX and BRD4 interactions in A549 cells treated with Garcinol. Red foci indicate close proximity of the two proteins. Blank arrows indicate interactions of ISX with BRD4 in the cells. NC: negative control.
- G
Western blotting analysis of the protein levels of ISX, PCAF, and EMT markers in A549 cells with PCAF knockdown and DOX‐inducible ISX expression system at 8 h time point after DOX induction.
- H, I
Cell migration (wound healing, h) and invasion (Transwell, I) assay of A549 lung cancer cells treated with Garcinol. V, vehicle. Data are presented as mean ± SD in graph (***P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate. Scale bar, 100 μm.

- A, B
Schematic representation of the potential acetylation domain organization of ISX and its lysine mutants (AC1–AC3).
- C
Recombinant PCAF acetylates His6‐ISX at lysine residue 69 by in vitro acetylation assay. Acetylated ISX was detected by anti‐acetyl Lysine antibody.
- D, E
The protein levels of GFP‐tagged WT or mutant ISX, PCAF, and BRD4 were determined in cytosol, nuclei, and anti‐GFP immunoprecipitates of A549 cells by Western blotting. Acetylated ISX was detected by anti‐acetyl Lysine antibody.
- F
The protein levels of total and acetylated histone H3 were determined in anti‐histone H3 immunoprecipitates of A549 cells by Western blotting.
- G, H
The cell migration (wound healing, G) and invasion (Transwell, H) activity were determined in A549 cells with GFP‐tagged wild or ISX mutants. Data are presented as mean ± SD in graph (***P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate. Scale bar, 100 μm.
- I
Schematic representation of tumor xenograft metastasis activity of constitutively expressing RFP A549 cells transfected with wild‐type or ISX AC3 mutant cDNA. Modified A549 cells were directly injected into lung to form xenograft tumor mass.
- J
Tumor xenograft metastasis activity of constitutively expressing RFP A549 cells transfected with wild‐type or ISX AC3 mutant cDNA was imaged by IVIS imaging system at fifth weeks.
- K
Kaplan–Meier survival curve analysis of nude mice xenograft injected with A549 cells carrying GFP, ISX‐GFP, and ISX Ac‐GFP (AC3) (n = 10). P = 0.0003. P‐values were calculated by log‐rank (Mantel–Cox) test comparing the two Kaplan–Meier curves.
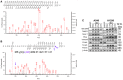
- A, B
PCAF acetylation of ISX was detected at lysine residue 69 by liquid chromatography–tandem mass spectrometry (LC‐tandem MS‐MS). The peptide NH2‐SDMDRPEGPGEEGPGEAAASGSGLEKPPK‐COOH of ISX (amino acids 44–72 a.a.) was identified with acetylated lysine at position 69 (y4, arrow).
- C
Expression levels of EMT markers were detected in A549 and H1299 cells transfected with wild‐type and mutants of ISX cDNA by Western blotting.

- A, B
Schematic representation of the potential acetylation domain organization of BRD4 and its lysine mutants (AC1–AC4).
- C
Recombinant PCAF acetylates His6‐BRD4 at lysine residue 332. Acetylated BRD4 was detected by anti‐acetyl lysine antibody.
- D
The mCherry‐tagged WT and BRD4 mutants were detected in anti‐GFP immunoprecipitates by Western blotting in A549 cells.
- E, F
The mRNA levels of TWIST1(E) and Snail1(F) were verified in A549 cells co‐expressing mCherry‐tagged WT or mutant BRD4 and GFP‐tagged ISX by RT–PCR. Data are presented as mean ± SD in bar graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- G, H
The DNA‐binding activity of TWIST1 and Snail1 was evaluated in anti‐GFP‐mCherry ChIP–ChIP immunoprecipitates by RT–PCR in A549 cells. Red, Hprt1 promoter (−190/+40 bp, negative control) (10% input of each group was pulled down and used for qPCR analysis). Data are presented as mean ± SD in bar graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- I
The cell invasion (Transwell) activity was determined in A549 cells co‐transfected with cDNA coding for GFP‐tagged ISX and mCherry‐tagged BRD4 mutants. Data are presented as mean ± SD in bar graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
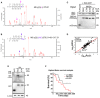
- A, B
PCAF acetylation of BRD4 is detected at lysine residue 332 by liquid chromatography–tandem mass spectrometry (LC‐tandem MS‐MS). The peptide NH2‐ESSRPVKPPKK‐COOH of BRD4 (amino acids 323–333 a.a.) was identified with acetylation lysine acetylated at position 332 (y(2), arrow).
- C
Ectopic expression of ISX‐GFP and wild‐type and mutants of BRD4 was detected by Western blotting in A549 cells.
- D
Correlation between BRD4 and PCAF mRNA levels detected by real‐time PCR in 157 lung cancer samples.
- E
The antibody generated by acetylated ISX‐K69 peptide detects GFP‐tagged wild‐type ISX, but not GFP‐tagged ISX‐K69 mutant.
- F
The Kaplan–Meier survival curve was used to analyze survival correlation between patients with NSCLC (n = 157) and BRD4. On the basis of the cut‐off values of fold differences, the study population was dichotomized into “high” and “low” expression groups. P‐values were calculated by log‐rank (Mantel–Cox) test comparing the two Kaplan–Meier curves.

- A
Schematic depiction of ISX and deletion constructs used.
- B
GFP‐tagged truncated ISX mutant proteins or HA‐tagged BRD4 was detected in anti‐HA or GFP immunoprecipitates by Western blotting analysis.
- C–F
A putative interaction surface of the ISX/BRD4 complex was presented through structural modeling. The electrostatic surfaces are drawn either blue for positive or red for negative charge (E and F).

- A
mCherry‐tagged truncated BRD4 mutant proteins and GFP‐tagged ISX were detected in anti‐GFP immunoprecipitates by Western blot.
- B, C
The DNA‐binding activity of Snail (B) and TWIST1 (C) was evaluated in anti‐GFP‐mCherry ChIP–ChIP immunoprecipitates by RT–PCR in A549 cells. Red, Hprt1 promoter (−190 to +40 bp, negative control). (10% input of each group was pull down and applied to qPCR) Data are presented as mean ± SD in bar graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.
- D, E
The cell migration (wound healing, D) and invasion (Transwell, E) activity were determined in A549 cells co‐transfected with cDNAs for GFP‐tagged ISX and mCherry‐tagged BRD4 mutants. Data are presented as mean ± SD in bar graph (P < 0.001, Student's t‐test) of three independent experiments, each performed in triplicate.

- A
IHC staining with ISX (brown) or PCAF (brown) antibody in lung tumors from patients with lung cancer. N, normal tissue; T, tumor mass; Nuclei, hematoxylin (blue); Blue and red arrow, cytosol and nuclear ISX (PCAF). Scale bar, 500 μm (400×); Scale bar, 2 mm (100×).
- B
Confocal immunofluorescence detection of ISX (green) and BRD4 (red) in lung tumors from patients with NSCLC. Cell nuclei were visualized by DAPI (blue); yellow arrows indicate co‐localization; IgG, negative control. N, normal tissue; T, tumor mass.
- C
Confocal immunofluorescence detection of ISX (green) and PCAF (red) in lung tumors from patients with NSCLC. Cell nuclei were visualized by DAPI (blue); yellow arrows indicate co‐localization; IgG, negative control. N, normal tissue; T, tumor mass.
- D, E
Correlation analysis of mRNA expression for, (D) ISX and BRD4, (E) ISX and PCAF in 157 lung cancer samples.
- F
Protein expression of ISX, ISX(69Kac), and vimentin in tumor and normal parts from patients with NSCLC by Western blotting. N, normal tissue; T, tumor mass; Arrow, target protein; *, unspecific band.
- G, H
The Kaplan–Meier survival curve was used to analyze survival correlation between patients (n = 157) with NSCLC and ISX (G) and PCAF (H) levels. On the basis of the cut‐off values of fold differences, the study population was dichotomized into the “high” and “low” expression groups. P‐values were calculated by log‐rank (Mantel–Cox) test comparing the two Kaplan–Meier curves.

References
-
- Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: a landscape takes shape. Cell 128: 635–638 - PubMed
-
- Schlesinger S, Meshorer E (2019) Open chromatin, epigenetic plasticity, and nuclear organization in pluripotency. Dev Cell 48: 135–150 - PubMed
-
- Jones PA, Ohtani H, Chakravarthy A, De Carvalho DD (2019) Epigenetic therapy in immune‐oncology. Nat Rev Cancer 19: 151–161 - PubMed
-
- Cheng Z, Lei Z, Yang P, Si A, Xiang D, Tang X, Guo G, Zhou J, Huser N (2019) Exosome‐transmitted p120‐catenin suppresses hepatocellular carcinoma progression via STAT3 pathways. Mol Carcinog 58: 1389–1399 - PubMed
-
- Hamamori Y, Sartorelli V, Ogryzko V, Puri PL, Wu HY, Wang JY, Nakatani Y, Kedes L (1999) Regulation of histone acetyltransferases p300 and PCAF by the bHLH protein twist and adenoviral oncoprotein E1A. Cell 96: 405–413 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- EOPP10-014/National Health Research Institutes (NHRI)/International
- MOST-105-2314-B-037-70-MY2/Ministry of Science and Technology, Taiwan (MOST)/International
- MOST-106-2314-B-037-090-MY2/Ministry of Science and Technology, Taiwan (MOST)/International
- KMU-TC108A04-5/Kaohsiung Medical University Chung-Ho Memorial Hospital (KMUH)/International
- MOST-107-2314-B-037-028-MY3/Ministry of Science and Technology, Taiwan (MOST)/International
- KMUH107-7R34/Kaohsiung Medical University Chung-Ho Memorial Hospital (KMUH)/International
- KMUH107-7R46/Kaohsiung Medical University Chung-Ho Memorial Hospital (KMUH)/International
- R01 CA251698/CA/NCI NIH HHS/United States
- MOST-108-2320-B-037-005/Ministry of Science and Technology, Taiwan (MOST)/International
- 106KMUOR04/Kaohsiung Medical University (KMU)/International
- MOST-107-2314-B-037-063-MY3/Ministry of Science and Technology, Taiwan (MOST)/International
- KQTD20170331145453160/Shenzhen Science and Technology Peacock Team Project/International
- RP190077/Cancer Prevention & Research Institute of Texas/International
- I-1805/Welch Foundation/International
- KMU-TC108A02/Kaohsiung Medical University (KMU)/International
- MOST-108-2314-B-037-065-MY3/Ministry of Science and Technology, Taiwan (MOST)/International
- KMU-DK108012/Kaohsiung Medical University (KMU)/International
- KMUH-106-6R33/Kaohsiung Medical University Chung-Ho Memorial Hospital (KMUH)/International
- NHRI-EX108-10720SI/National Health Research Institutes (NHRI)/International
- R01 CA103867/CA/NCI NIH HHS/United States
- CPRIT RP180349/Cancer Prevention & Research Institute of Texas/International
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

