Comprehensive multi-omics analysis identified core molecular processes in esophageal cancer and revealed GNGT2 as a potential prognostic marker
- PMID: 31908393
- PMCID: PMC6938725
- DOI: 10.3748/wjg.v25.i48.6890
Comprehensive multi-omics analysis identified core molecular processes in esophageal cancer and revealed GNGT2 as a potential prognostic marker
Abstract
Background: Esophageal cancer is one of the most poorly diagnosed and fatal cancers in the world. Although a series of studies on esophageal cancer have been reported, the molecular pathogenesis of the disease remains elusive.
Aim: To investigate comprehensively the molecular process of esophageal cancer.
Methods: Differential expression analysis was performed to identify differentially expressed genes (DEGs) in different stages of esophageal cancer from The Cancer Genome Atlas data. Exacting gene interaction modules were generated, and hub genes in the module interaction network were found. Further, through survival analysis, methylation analysis, pivot analysis, and enrichment analysis, some important molecules and related functions/pathways were identified to elucidate potential mechanisms in esophageal cancer.
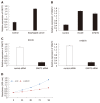
Results: A total of 7457 DEGs and 14 gene interaction modules were identified. These module genes were significantly involved in the positive regulation of protein transport, gastric acid secretion, insulin-like growth factor receptor binding, and other biological processes as well as p53 signaling pathway, epidermal growth factor signaling pathway, and epidermal growth factor receptor signaling pathway. Transcription factors (including hypoxia inducible factor 1A) and non-coding RNAs (including colorectal differentially expressed and hsa-miR-330-3p) that significantly regulate dysfunction modules were identified. Survival analysis showed that G protein subunit gamma transducin 2 (GNGT2) was closely related to survival of esophageal cancer. DEGs with strong methylation regulation ability were identified, including SST and SH3GL2. Furthermore, the expression of GNGT2 was evaluated by quantitative real time polymerase chain reaction, and the results showed that GNGT2 expression was significantly upregulated in esophageal cancer patient samples and cell lines. Moreover, cell counting kit-8 assay revealed that GNGT2 could promote the proliferation of esophageal cancer cell lines.
Conclusion: This study not only revealed the potential regulatory factors involved in the development of esophageal cancer but also deepens our understanding of its underlying mechanism.
Keywords: Enrichment analysis; Esophageal cancer; GNGT2; Gene interaction module; Molecular pathogenesis; Regulatory factors.
©The Author(s) 2019. Published by Baishideng Publishing Group Inc. All rights reserved.
Conflict of interest statement
Conflict-of-interest statement: The authors declare no conflict of interest.
Figures


References
-
- Yang DH, Jung HY. [Treatment of esophageal cancer] Korean J Gastroenterol. 2008;52(6):338–350. - PubMed
-
- Li HY, Liu YC, Bai YH, Sun M, Wang L, Zhang XB, Cai B. SNP at miR-483-5p-binding site in the 3'-untranslated region of the BSG gene is associated with susceptibility to esophageal cancer in a Chinese population. Genet Mol Res. 2016;(6):15(2). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

