Evaluation of freely available software tools for untargeted quantification of 13C isotopic enrichment in cellular metabolome from HR-LC/MS data
- PMID: 31908925
- PMCID: PMC6940703
- DOI: 10.1016/j.mec.2019.e00120
Evaluation of freely available software tools for untargeted quantification of 13C isotopic enrichment in cellular metabolome from HR-LC/MS data
Erratum in
-
Erratum regarding previously published articles in volumes 9, 10 and 11.Metab Eng Commun. 2021 Oct 28;13:e00186. doi: 10.1016/j.mec.2021.e00186. eCollection 2021 Dec. Metab Eng Commun. 2021. PMID: 34765440 Free PMC article.
Abstract
13C Metabolic Flux Analysis (13C-MFA) involves the quantification of isotopic enrichment in cellular metabolites and fitting the resultant data to the metabolic network model of the organism. Coverage and resolution of the resultant flux map depends on the total number of metabolites and fragments in which 13C enrichment can be quantified accurately. Experimental techniques for tracking 13C enrichment are evolving rapidly and large volumes of data are now routinely generated through the use of Liquid Chromatography coupled with High-Resolution Mass Spectrometry (HR-LC/MS). Therefore, the current manuscript is focused on the challenges in high-throughput analyses of such large datasets. Current 13C-MFA studies often have to rely on the targeted quantification of a small subset of metabolites, thereby leaving a large fraction of the data unexplored. A number of public domain software tools have been reported in recent years for the untargeted quantitation of isotopic enrichment. However, the suitability of their application across diverse datasets has not been investigated. Here, we test the software tools X13CMS, DynaMet, geoRge, and HiResTEC with three diverse datasets. The tools provided a global, untargeted view of 13C enrichment in metabolites in all three datasets and a much-needed automation in data analysis. Some inconsistencies were observed in results obtained from the different tools, which could be partially ascribed to the lack of baseline separation and potential mass conflicts. After removing the false positives manually, isotopic enrichment could be quantified reliably in a large repertoire of metabolites. Of the software tools explored, geoRge and HiResTEC consistently performed well for the untargeted analysis of all datasets tested.
Keywords: 13C metabolic flux analysis; Cyanobacteria; Methanolicus; Reticulocytes; Synechococcus sp. PCC 7002; Untargeted analysis.
© 2019 The Authors.
Figures
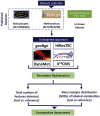
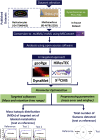
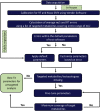
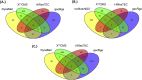





References
-
- Alagesan S., Gaudana S.B., Sinha A., Wangikar P.P. Metabolic flux analysis of Cyanothece sp. ATCC 51142 under mixotrophic conditions. Photosynth. Res. 2013;118:191–198. - PubMed
-
- Capellades J., Navarro M., Samino S., Garcia-Ramirez M., Hernandez C., Simo R., Vinaixa M., Yanes O. GeoRge: a computational tool to detect the presence of stable isotope labeling in LC/MS-based untargeted metabolomics. Anal. Chem. 2016;88:621–628. - PubMed
-
- Choi J., Antoniewicz M.R. Tandem mass spectrometry: a novel approach for metabolic flux analysis. Metab. Eng. 2011;13:225–233. - PubMed
LinkOut - more resources
Full Text Sources

