Profiling of lincRNAs in human pluripotent stem cell derived forebrain neural progenitor cells
- PMID: 31909251
- PMCID: PMC6940631
- DOI: 10.1016/j.heliyon.2019.e03067
Profiling of lincRNAs in human pluripotent stem cell derived forebrain neural progenitor cells
Abstract
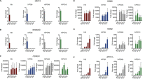
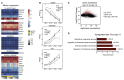
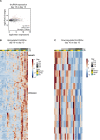
Human embryonic stem cells (hESCs) and induced pluripotent stem cells (iPSCs) can be differentiated into many different cell types of the central nervous system. One challenge when using pluripotent stem cells is to develop robust and efficient differentiation protocols that result in homogenous cultures of the desired cell type. Here, we have utilized the SMAD-inhibitors SB431542 and Noggin in a fully defined monolayer culture model to differentiate human pluripotent cells into homogenous forebrain neural progenitors. Temporal fate analysis revealed that this protocol results in forebrain-patterned neural progenitor cells that start to express early neuronal markers after two weeks of differentiation, allowing for the analysis of gene expression changes during neurogenesis. Using this system, we were able to identify many previously uncharacterized long intergenic non-coding RNAs that display dynamic expression during human forebrain neurogenesis.
Keywords: Cell biology; Cellular neuroscience; Developmental genetics; Differentiation; Forebrain development; Genetics; Induced pluripotent stem cells; Neural progenitor cells; Neuroscience; lincRNAs.
© 2019 The Author(s).
Figures

References
-
- Campbell K. Dorsal-ventral patterning in the mammalian telencephalon. Curr. Opin. Neurobiol. 2003;13:50–56. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

