DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot
- PMID: 31910327
- PMCID: PMC7292547
- DOI: 10.1111/pbi.13325
DcMYB113, a root-specific R2R3-MYB, conditions anthocyanin biosynthesis and modification in carrot
Abstract
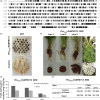
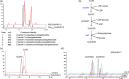
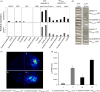
Purple carrots, the original domesticated carrots, accumulate highly glycosylated and acylated anthocyanins in root and/or petiole. Previously, a quantitative trait locus (QTL) for root-specific anthocyanin pigmentation was genetically mapped to chromosome 3 of carrot. In this study, an R2R3-MYB gene, namely DcMYB113, was identified within this QTL region. DcMYB113 expressed in the root of 'Purple haze', a carrot cultivar with purple root and nonpurple petiole, but not in the roots of two carrot cultivars with a purple root and petiole (Deep purple and Cosmic purple) and orange carrot 'Kurodagosun', which appeared to be caused by variation in the promoter region. The function of DcMYB113 from 'Purple haze' was verified by transformation in 'Cosmic purple' and 'Kurodagosun', resulting in anthocyanin biosynthesis. Transgenic 'Kurodagosun' carrying DcMYB113 driven by the CaMV 35S promoter had a purple root and petiole, while transgenic 'Kurodagosun' expressing DcMYB113 driven by its own promoter had a purple root and nonpurple petiole, suggesting that root-specific expression of DcMYB113 was determined by its promoter. DcMYB113 could activate the expression of DcbHLH3 and structural genes related to anthocyanin biosynthesis. DcUCGXT1 and DcSAT1, which were confirmed to be responsible for anthocyanins glycosylation and acylation, respectively, were also activated by DcMYB113. The WGCNA identified several genes co-expressed with anthocyanin biosynthesis and the results indicated that DcMYB113 may regulate anthocyanin transport. Our findings provide insight into the molecular mechanism underlying root-specific anthocyanin biosynthesis and further modification in carrot and even other root crops.
Keywords: DcMYB113; WGCNA; anthocyanin; carrot; modification; root-specific.
© 2020 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
All co‐authors declared that they have no conflicts of interest in this work.
Figures




References
-
- Arscott, S.A. and Tanumihardjo, S.A. (2010) Carrots of many colors provide basic nutrition and bioavailable phytochemicals acting as a functional food. Compr. Rev. Food Sci. Food Saf. 9, 223–239.
-
- Banga, O. (1963) Origin and distribution of the western cultivated carrot. Genet. Agrar. 17, 357–370.
-
- Baranski, R. , Maksylewicz‐Kaul, A. , Nothnagel, T. , Cavagnaro, P.F. , Simon, P.W. and Grzebelus, D. (2012) Genetic diversity of carrot (Daucus carota L.) cultivars revealed by analysis of SSR loci. Genet. Resour. Crop Evol. 59, 163–170.
-
- Butelli, E. , Titta, L. , Giorgio, M. , Mock, H.P. , Matros, A. , Peterek, S. , Schijlen, E.G.W.M. et al. (2008) Enrichment of tomato fruit with health‐promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

