Evolutionary rates are correlated between cockroach symbionts and mitochondrial genomes
- PMID: 31910734
- PMCID: PMC7013487
- DOI: 10.1098/rsbl.2019.0702
Evolutionary rates are correlated between cockroach symbionts and mitochondrial genomes
Abstract
Bacterial endosymbionts evolve under strong host-driven selection. Factors influencing host evolution might affect symbionts in similar ways, potentially leading to correlations between the molecular evolutionary rates of hosts and symbionts. Although there is evidence of rate correlations between mitochondrial and nuclear genes, similar investigations of hosts and symbionts are lacking. Here, we demonstrate a correlation in molecular rates between the genomes of an endosymbiont (Blattabacterium cuenoti) and the mitochondrial genomes of their hosts (cockroaches). We used partial genome data for multiple strains of B. cuenoti to compare phylogenetic relationships and evolutionary rates for 55 cockroach/symbiont pairs. The phylogenies inferred for B. cuenoti and the mitochondrial genomes of their hosts were largely congruent, as expected from their identical maternal and cytoplasmic mode of inheritance. We found a correlation between evolutionary rates of the two genomes, based on comparisons of root-to-tip distances and on comparisons of the branch lengths of phylogenetically independent species pairs. Our results underscore the profound effects that long-term symbiosis can have on the biology of each symbiotic partner.
Keywords: Blattabacterium cuenoti; cockroach; host–symbiont interaction; molecular evolution; phylogeny; substitution rate.
Conflict of interest statement
We declare we have no competing interests.
Figures
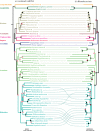
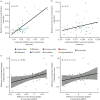
References
-
- Ho SYW, Lo N. 2013. The insect molecular clock. Aust. J. Entomol. 52, 101–105. (10.1111/aen.12018) - DOI
-
- Douglas AE. 2010. The symbiotic habit. Princeton, NJ: Princeton University Press.
Publication types
MeSH terms
Associated data
LinkOut - more resources
Full Text Sources
