Phylogenetic analysis of porcine circovirus type 2 (PCV2) between 2015 and 2018 in Henan Province, China
- PMID: 31910824
- PMCID: PMC6947828
- DOI: 10.1186/s12917-019-2193-1
Phylogenetic analysis of porcine circovirus type 2 (PCV2) between 2015 and 2018 in Henan Province, China
Abstract
Background: Porcine circovirus type 2 (PCV2) is the pathogen of porcine circovirus associated diseases (PCVAD) and one of the main pathogens in the global pig industry, which has brought huge economic losses to the pig industry. In recent years, there has been limited research on the prevalence of PCV2 in Henan Province. This study investigated the genotype and evolution of PCV2 in this area.
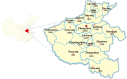
Results: We collected 117 clinical samples from different regions of Henan Province from 2015 to 2018. Here, we found that the PCV2 infection rate of PCV2 was 62.4%. Thirty-seven positive clinical samples were selected to amplify the complete genome of PCV2 and were sequenced. Based on the phylogenetic analysis of PCV2 ORF2 and complete genome, it was found that the 37 newly detected strains belonged to PCV2a (3 of 37), PCV2b (21 of 37) and PCV2d (13 of 37), indicating the predominant prevalence of PCV2b and PCV2d strains. In addition, we compared the amino acid sequences and found several amino acid mutation sites among different genotypes. Furthermore, the results of selective pressure analysis showed that there were 5 positive selection sites.
Conclusions: This study indicated the genetic diversity, molecular epidemiology and evolution of PCV2 genotypes in Henan Province during 2015-2018.
Keywords: Evolution; Genetic diversity; Molecular epidemiology; PCV2; Phylogenetic analysis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

References
MeSH terms
LinkOut - more resources
Full Text Sources

