Multi-omic serum biomarkers for prognosis of disease progression in prostate cancer
- PMID: 31910880
- PMCID: PMC6945688
- DOI: 10.1186/s12967-019-02185-y
Multi-omic serum biomarkers for prognosis of disease progression in prostate cancer
Abstract
Background: Predicting the clinical course of prostate cancer is challenging due to the wide biological spectrum of the disease. The objective of our study was to identify prostate cancer prognostic markers in patients 'sera using a multi-omics discovery platform.
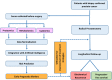
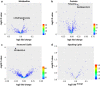
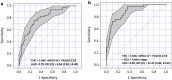
Methods: Pre-surgical serum samples collected from a longitudinal, racially diverse, prostate cancer patient cohort (N = 382) were examined. Linear Regression and Bayesian computational approaches integrated with multi-omics, were used to select markers to predict biochemical recurrence (BCR). BCR-free survival was modeled using unadjusted Kaplan-Meier estimation curves and multivariable Cox proportional hazards analysis, adjusted for key pathologic variables. Receiver operating characteristic (ROC) curve statistics were used to examine the predictive value of markers in discriminating BCR events from non-events. The findings were further validated by creating a training set (N = 267) and testing set (N = 115) from the cohort.
Results: Among 382 patients, 72 (19%) experienced a BCR event in a median follow-up time of 6.9 years. Two proteins-Tenascin C (TNC) and Apolipoprotein A1V (Apo-AIV), one metabolite-1-Methyladenosine (1-MA) and one phospholipid molecular species phosphatidic acid (PA) 18:0-22:0 showed a cumulative predictive performance of AUC = 0.78 [OR (95% CI) = 6.56 (2.98-14.40), P < 0.05], in differentiating patients with and without BCR event. In the validation set all four metabolites consistently reproduced an equivalent performance with high negative predictive value (NPV; > 80%) for BCR. The combination of pTstage and Gleason score with the analytes, further increased the sensitivity [AUC = 0.89, 95% (CI) = 4.45-32.05, P < 0.05], with an increased NPV (0.96) and OR (12.4) for BCR. The panel of markers combined with the pathological parameters demonstrated a more accurate prediction of BCR than the pathological parameters alone in prostate cancer.
Conclusions: In this study, a panel of serum analytes were identified that complemented pathologic patient features in predicting prostate cancer progression. This panel offers a new opportunity to complement current prognostic markers and to monitor the potential impact of primary treatment versus surveillance on patient oncological outcome.
Keywords: Bayesian networks; Biochemical recurrence; Biomarkers; Metabolomics; Prostate cancer.
Conflict of interest statement
MAK, NRN, RS, VA, LOR, LZ, SS, AD, and JC are co-inventors on patent application “Markers for the Diagnosis of Prostate Cancer”. Other authors declare that they have no competing interests.
MAK, EM, LOR, EYC, VT, LZ, KP, PS, EG, RS, VA, and NRN are current or former employees of BERG, LLC and have stock options. NRN is co-founder of BERG, LLC.
Figures

References
-
- Cooperberg MR, Davicioni E, Crisan A, Jenkins RB, Ghadessi M, Karnes RJ. Combined value of validated clinical and genomic risk stratification tools for predicting prostate cancer mortality in a high-risk prostatectomy cohort. Eur Urol. 2015;67(2):326–333. doi: 10.1016/j.eururo.2014.05.039. - DOI - PMC - PubMed
-
- Cullen J, Rosner IL, Brand TC, Zhang N, Tsiatis AC, Moncur J, et al. A Biopsy-based 17-gene genomic prostate score predicts recurrence after radical prostatectomy and adverse surgical pathology in a racially diverse population of men with clinically low- and intermediate-risk prostate cancer. Eur Urol. 2015;68(1):123–131. doi: 10.1016/j.eururo.2014.11.030. - DOI - PubMed
-
- Klein EA, Cooperberg MR, Magi-Galluzzi C, Simko JP, Falzarano SM, Maddala T, et al. A 17-gene assay to predict prostate cancer aggressiveness in the context of Gleason grade heterogeneity, tumor multifocality, and biopsy undersampling. Eur Urol. 2014;66(3):550–560. doi: 10.1016/j.eururo.2014.05.004. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

