Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10
- PMID: 31911601
- PMCID: PMC6946704
- DOI: 10.1038/s41467-019-13936-2
Hand-foot-and-mouth disease virus receptor KREMEN1 binds the canyon of Coxsackie Virus A10
Abstract
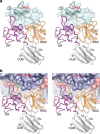
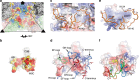
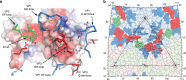
Coxsackievirus A10 (CV-A10) is responsible for an escalating number of severe infections in children, but no prophylactics or therapeutics are currently available. KREMEN1 (KRM1) is the entry receptor for the largest receptor-group of hand-foot-and-mouth disease causing viruses, which includes CV-A10. We report here structures of CV-A10 mature virus alone and in complex with KRM1 as well as of the CV-A10 A-particle. The receptor spans the viral canyon with a large footprint on the virus surface. The footprint has some overlap with that seen for the neonatal Fc receptor complexed with enterovirus E6 but is larger and distinct from that of another enterovirus receptor SCARB2. Reduced occupancy of a particle-stabilising pocket factor in the complexed virus and the presence of both unbound and expanded virus particles suggests receptor binding initiates a cascade of conformational changes that produces expanded particles primed for viral uncoating.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Molecular basis of Coxsackievirus A10 entry using the two-in-one attachment and uncoating receptor KRM1.Proc Natl Acad Sci U S A. 2020 Aug 4;117(31):18711-18718. doi: 10.1073/pnas.2005341117. Epub 2020 Jul 20. Proc Natl Acad Sci U S A. 2020. PMID: 32690697 Free PMC article.
-
KREMEN1 Is a Host Entry Receptor for a Major Group of Enteroviruses.Cell Host Microbe. 2018 May 9;23(5):636-643.e5. doi: 10.1016/j.chom.2018.03.019. Epub 2018 Apr 19. Cell Host Microbe. 2018. PMID: 29681460
-
Completely conserved VP2 residue K140 of KREMEN1-dependent enteroviruses is critical for virus-receptor interactions and viral infection.mBio. 2025 Feb 5;16(2):e0304024. doi: 10.1128/mbio.03040-24. Epub 2025 Jan 16. mBio. 2025. PMID: 39817751 Free PMC article.
-
Cellular receptors for enterovirus A71.J Biomed Sci. 2020 Jan 10;27(1):23. doi: 10.1186/s12929-020-0615-9. J Biomed Sci. 2020. PMID: 31924205 Free PMC article. Review.
-
Hand, foot, and mouth disease associated with coxsackievirus A10: more serious than it seems.Expert Rev Anti Infect Ther. 2019 Apr;17(4):233-242. doi: 10.1080/14787210.2019.1585242. Epub 2019 Mar 6. Expert Rev Anti Infect Ther. 2019. PMID: 30793637 Review.
Cited by
-
Atomic Structures of Coxsackievirus B5 Provide Key Information on Viral Evolution and Survival.J Virol. 2022 May 11;96(9):e0010522. doi: 10.1128/jvi.00105-22. Epub 2022 Apr 20. J Virol. 2022. PMID: 35442060 Free PMC article.
-
Identification of Critical Amino Acids of Coxsackievirus A10 Associated with Cell Tropism and Viral RNA Release during Uncoating.Viruses. 2023 Oct 18;15(10):2114. doi: 10.3390/v15102114. Viruses. 2023. PMID: 37896891 Free PMC article.
-
Identification of a neutralizing linear epitope within the VP1 protein of coxsackievirus A10.Virol J. 2022 Dec 1;19(1):203. doi: 10.1186/s12985-022-01939-3. Virol J. 2022. PMID: 36457099 Free PMC article.
-
Serotype specific epitopes identified by neutralizing antibodies underpin immunogenic differences in Enterovirus B.Nat Commun. 2020 Sep 4;11(1):4419. doi: 10.1038/s41467-020-18250-w. Nat Commun. 2020. PMID: 32887892 Free PMC article.
-
'Tomato flu' a new epidemic in India: Virology, epidemiology, and clinical features.New Microbes New Infect. 2022 Dec 13;51:101070. doi: 10.1016/j.nmni.2022.101070. eCollection 2023 Jan. New Microbes New Infect. 2022. PMID: 36582550 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

