Up-regulation of circ_LARP4 suppresses cell proliferation and migration in ovarian cancer by regulating miR-513b-5p/LARP4 axis
- PMID: 31911757
- PMCID: PMC6945592
- DOI: 10.1186/s12935-019-1071-z
Up-regulation of circ_LARP4 suppresses cell proliferation and migration in ovarian cancer by regulating miR-513b-5p/LARP4 axis
Abstract
Background: Ovarian cancer (OC) is a common fatal malignant tumor of female reproductive system worldwide. Growing studies have proofed that circular RNAs (circRNAs) engage in the regulation of various types of cancers. However, the underlying biological functions and effect mechanism of circular RNA_LARP4 (circ_LARP4) in OC have not been explored.
Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) analysis was used to detect the expression of circ_LARP4 in OC cells. The function of circ_LARP4 was measured by cell counting kit-8 (CCK-8), colony formation assay and transwell assay. RNA immunoprecipitation (RIP) assay and luciferase reporter assays assessed the binding correlation between miR-513b-5p and circ_LARP4 (or LARP4).
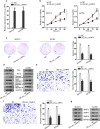
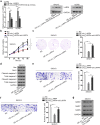
Results: The expression of circ_LARP4 in OC cells was much lower than that in human normal ovarian epithelial cells. Overexpressing circ_LARP4 impaired cell proliferation, invasion and migration abilities. Circ_LARP4 worked as a competing endogenous RNA (ceRNA) to sponge miR-513b-5p. Furthermore, LARP4 was indirectly modulated by circ_LARP4 as the downstream target of miR-513b-5p, as well as the host gene of circ_LARP4.
Conclusion: Circ_LARP4 could hamper cell proliferation and migration by sponging miR-513b-5p to regulate the expression of LARP4. This research may provide some referential value to OC treatment.
Keywords: LARP4; OC; ceRNA; circ_LARP4; miR-513b-5p.
© The Author(s) 2020.
Conflict of interest statement
Competing interestsThe authors declare that they have no competing interests.
Figures



Similar articles
-
Hsa_circ_0018909 promotes non-small cell lung cancer by directly regulating hsa-miR-513b-5p.Front Oncol. 2025 Jun 23;15:1542742. doi: 10.3389/fonc.2025.1542742. eCollection 2025. Front Oncol. 2025. PMID: 40626007 Free PMC article.
-
Circular RNA Circ_0025033 Promotes the Evolvement of Ovarian Cancer Through the Regulation of miR-330-5p/KLK4 Axis.Cancer Manag Res. 2020 Apr 23;12:2753-2765. doi: 10.2147/CMAR.S241372. eCollection 2020. Cancer Manag Res. 2020. Retraction in: Cancer Manag Res. 2021 Dec 23;13:9345-9346. doi: 10.2147/CMAR.S354552. PMID: 32425594 Free PMC article. Retracted.
-
Circular RNA La-Related Protein 4 Inhibits Non-Small Cell Lung Cancer Cell Proliferation While Promotes Apoptosis Through Sponging microRNA-21-5p.Cancer Biother Radiopharm. 2022 Mar;37(2):111-118. doi: 10.1089/cbr.2020.3707. Epub 2020 Jul 2. Cancer Biother Radiopharm. 2022. PMID: 32614609
-
Effect of Solanum lyratum Polysaccharide on Malignant Behaviors of Lung Cancer Cells by Regulating the Circ_UHRF1/miR-513b-5p Axis.Cell Mol Biol (Noisy-le-grand). 2022 Feb 27;67(6):191-199. doi: 10.14715/cmb/2021.67.6.26. Cell Mol Biol (Noisy-le-grand). 2022. PMID: 35818196
-
Circ_0015756 promotes the progression of ovarian cancer by regulating miR-942-5p/CUL4B pathway.Cancer Cell Int. 2020 Nov 27;20(1):572. doi: 10.1186/s12935-020-01666-1. Cancer Cell Int. 2020. PMID: 33292255 Free PMC article.
Cited by
-
CircKIAA0907 Retards Cell Growth, Cell Cycle, and Autophagy of Gastric Cancer In Vitro and Inhibits Tumorigenesis In Vivo via the miR-452-5p/KAT6B Axis.Med Sci Monit. 2020 Jul 28;26:e924160. doi: 10.12659/MSM.924160. Med Sci Monit. 2020. PMID: 32722658 Free PMC article.
-
Hsa_circ_0063804 enhances ovarian cancer cells proliferation and resistance to cisplatin by targeting miR-1276/CLU axis.Aging (Albany NY). 2022 Jun 10;14(11):4699-4713. doi: 10.18632/aging.203474. Epub 2022 Jun 10. Aging (Albany NY). 2022. PMID: 35687899 Free PMC article.
-
N6-Methyladenosine-regulated LINC00675 suppress the proliferation, migration and invasion of breast cancer cells via inhibiting miR-513b-5p.Bioengineered. 2021 Dec;12(2):10690-10702. doi: 10.1080/21655979.2021.2001905. Bioengineered. 2021. PMID: 34738869 Free PMC article.
-
Immunoregulatory role of exosomal circRNAs in the tumor microenvironment.Front Oncol. 2025 Feb 17;15:1453786. doi: 10.3389/fonc.2025.1453786. eCollection 2025. Front Oncol. 2025. PMID: 40034598 Free PMC article. Review.
-
Intracranial Aneurysm Biomarkers: A Convergence of Genetics, Inflammation, Oxidative Stress, and the Extracellular Matrix.Int J Mol Sci. 2025 Apr 2;26(7):3316. doi: 10.3390/ijms26073316. Int J Mol Sci. 2025. PMID: 40244203 Free PMC article. Review.
References
-
- Pisanic TR, 2nd, Cope LM, Lin SF, Yen TT, Athamanolap P, Asaka R, Nakayama K, Fader AN, Wang TH, Shih IM, et al. Methylomic analysis of ovarian cancers identifies tumor-specific alterations readily detectable in early precursor lesions. Clin Cancer Res. 2018;24(24):6536–6547. doi: 10.1158/1078-0432.CCR-18-1199. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

