Integrative network analysis identifies cell-specific trans regulators of m6A
- PMID: 31912146
- PMCID: PMC7038928
- DOI: 10.1093/nar/gkz1206
Integrative network analysis identifies cell-specific trans regulators of m6A
Abstract
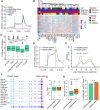
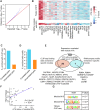
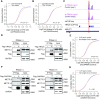
N6-methyladenosine (m6A) is a reversible and dynamic RNA modification in eukaryotes. However, how cells establish cell-specific m6A methylomes is still poorly understood. Here, we developed a computational framework to systematically identify cell-specific trans regulators of m6A through integrating gene expressions, binding targets and binding motifs of large number of RNA binding proteins (RBPs) with a co-methylation network constructed using large-scale m6A methylomes across diverse cell states. We applied the framework and successfully identified 32 high-confidence m6A regulators that modulated the variable m6A sites away from stop codons in a cell-specific manner. To validate them, we knocked down three regulators respectively and found two of them (TRA2A and CAPRIN1) selectively promoted the methylations of the m6A sites co-localized with their binding targets on RNAs through physical interactions with the m6A writers. Knockdown of TRA2A increased the stabilities of the RNAs with TRA2A bound near the m6A sites and decreased the viability of cells. The successful identification of m6A regulators demonstrates a powerful and widely applicable strategy to elucidate the cell-specific m6A regulators. Additionally, our discovery of pervasive trans-acting regulating of m6A provides novel insights into the mechanisms by which spatial and temporal dynamics of m6A methylomes are established.
© The Author(s) 2020. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures



References
-
- Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M. et al. .. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012; 485:201–206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

