A comprehensive global perspective on phylogenomics and evolutionary dynamics of Small ruminant morbillivirus
- PMID: 31913305
- PMCID: PMC6949297
- DOI: 10.1038/s41598-019-54714-w
A comprehensive global perspective on phylogenomics and evolutionary dynamics of Small ruminant morbillivirus
Erratum in
-
Author Correction: A comprehensive global perspective on phylogenomics and evolutionary dynamics of Small ruminant morbillivirus.Sci Rep. 2020 Nov 27;10(1):20997. doi: 10.1038/s41598-020-78219-z. Sci Rep. 2020. PMID: 33244107 Free PMC article.
Abstract
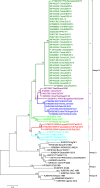
A string of complete genome sequences of Small ruminant morbillivirus (SRMV) have been reported from different parts of the globe including Asia, Africa and the Middle East. Despite individual genome sequence-based analysis, there is a paucity of comparative genomic and evolutionary analysis to provide overarching and comprehensive evolutionary insights. Therefore, we first enriched the existing database of complete genome sequences of SRMVs with Pakistan-originated strains and then explored overall nucleotide diversity, genomic and residue characteristics, and deduced an evolutionary relationship among strains representing a diverse geographical region worldwide. The average number of pairwise nucleotide differences among the whole genomes was found to be 788.690 with a diversity in nucleotide sequences (0.04889 ± S.D. 0.00468) and haplotype variance (0.00001). The RNA-dependent-RNA polymerase (L) gene revealed phylogenetic relationship among SRMVs in a pattern similar to those of complete genome and the nucleoprotein (N) gene. Therefore, we propose another useful molecular marker that may be employed for future epidemiological investigations. Based on evolutionary analysis, the mean evolution rate for the complete genome, N, P, M, F, H and L genes of SRMV was estimated to be 9.953 × 10-4, 1.1 × 10-3, 1.23 × 10-3, 2.56 × 10-3, 2.01 × 10-3, 1.47 × 10-3 and 9.75 × 10-4 substitutions per site per year, respectively. A recombinant event was observed in a Pakistan-originated strain (KY967608) revealing Indian strains as major (98.1%, KR140086) and minor parents (99.8%, KT860064). Taken together, outcomes of the study augment our knowledge and current understanding towards ongoing phylogenomic and evolutionary dynamics for better comprehensions of SRMVs and effective disease control interventions.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Genetic characterization of small ruminant morbillivirus from recently emerging wave of outbreaks in Pakistan.Transbound Emerg Dis. 2018 Dec;65(6):2032-2038. doi: 10.1111/tbed.12964. Epub 2018 Jul 26. Transbound Emerg Dis. 2018. PMID: 30051602
-
Genome sequencing of an Indian peste des petits ruminants virus isolate, Izatnagar/94, and its implications for virus diversity, divergence and phylogeography.Arch Virol. 2017 Jun;162(6):1677-1693. doi: 10.1007/s00705-017-3288-2. Epub 2017 Feb 28. Arch Virol. 2017. PMID: 28247095
-
Epidemic and evolutionary characteristics of peste des petits ruminants virus infecting Procapra przewalskii in Western China.Infect Genet Evol. 2019 Nov;75:104004. doi: 10.1016/j.meegid.2019.104004. Epub 2019 Aug 12. Infect Genet Evol. 2019. PMID: 31415822
-
Peste Des Petits Ruminants in Benin: Persistence of a Single Virus Genotype in the Country for Over 42 Years.Transbound Emerg Dis. 2017 Aug;64(4):1037-1044. doi: 10.1111/tbed.12471. Epub 2016 Jan 22. Transbound Emerg Dis. 2017. PMID: 26801518 Review.
-
Peste des petits ruminants in Africa: a review of currently available molecular epidemiological data, 2020.Arch Virol. 2020 Oct;165(10):2147-2163. doi: 10.1007/s00705-020-04732-1. Epub 2020 Jul 11. Arch Virol. 2020. PMID: 32653984 Free PMC article. Review.
Cited by
-
Peste des Petits Ruminants in Central and Eastern Asia/West Eurasia: Epidemiological Situation and Status of Control and Eradication Activities after the First Phase of the PPR Global Eradication Programme (2017-2021).Animals (Basel). 2022 Aug 10;12(16):2030. doi: 10.3390/ani12162030. Animals (Basel). 2022. PMID: 36009619 Free PMC article. Review.
-
Molecular Characterization of Lineage-IV Peste Des Petits Ruminants Virus and the Development of In-House Indirect Enzyme-Linked Immunosorbent Assay (IELISA) for its Rapid Detection".Biol Proced Online. 2024 Jul 5;26(1):22. doi: 10.1186/s12575-024-00249-y. Biol Proced Online. 2024. PMID: 38969986 Free PMC article.
-
Development of TaqMan-based real-time RT-PCR assay based on N gene for the quantitative detection of feline morbillivirus.BMC Vet Res. 2021 Mar 23;17(1):128. doi: 10.1186/s12917-021-02837-6. BMC Vet Res. 2021. PMID: 33757494 Free PMC article.
-
Immunoinformatics Approach for Epitope Mapping of Immunogenic Regions (N, F and H Gene) of Small Ruminant Morbillivirus and Its Comparative Analysis with Standard Vaccinal Strains for Effective Vaccine Development.Vaccines (Basel). 2022 Dec 19;10(12):2179. doi: 10.3390/vaccines10122179. Vaccines (Basel). 2022. PMID: 36560589 Free PMC article.
-
A curated dataset of peste des petits ruminants virus sequences for molecular epidemiological analyses.PLoS One. 2022 Feb 10;17(2):e0263616. doi: 10.1371/journal.pone.0263616. eCollection 2022. PLoS One. 2022. PMID: 35143560 Free PMC article.
References
-
- Munir M. ed. Peste des petits ruminants virus. Heidelberg, New York, Dordrecht, London: Springer. (2015).
MeSH terms
LinkOut - more resources
Full Text Sources

