Absence of GP130 cytokine receptor signaling causes extended Stüve-Wiedemann syndrome
- PMID: 31914175
- PMCID: PMC7062520
- DOI: 10.1084/jem.20191306
Absence of GP130 cytokine receptor signaling causes extended Stüve-Wiedemann syndrome
Abstract
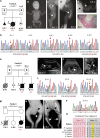
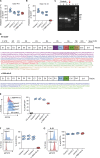
The gene IL6ST encodes GP130, the common signal transducer of the IL-6 cytokine family consisting of 10 cytokines. Previous studies have identified cytokine-selective IL6ST defects that preserve LIF signaling. We describe three unrelated families with at least five affected individuals who presented with lethal Stüve-Wiedemann-like syndrome characterized by skeletal dysplasia and neonatal lung dysfunction with additional features such as congenital thrombocytopenia, eczematoid dermatitis, renal abnormalities, and defective acute-phase response. We identified essential loss-of-function variants in IL6ST (a homozygous nonsense variant and a homozygous intronic splice variant with exon skipping). Functional tests showed absent cellular responses to GP130-dependent cytokines including IL-6, IL-11, IL-27, oncostatin M (OSM), and leukemia inhibitory factor (LIF). Genetic reconstitution of GP130 by lentiviral transduction in patient-derived cells reversed the signaling defect. This study identifies a new genetic syndrome caused by the complete lack of signaling of a whole family of GP130-dependent cytokines in humans and highlights the importance of the LIF signaling pathway in pre- and perinatal development.
© 2020 Chen et al.
Conflict of interest statement
Disclosures: Dr. Chen reported grants from Celgene during the conduct of the study and grants from Celgene outside the submitted work. Dr. Devey is an employee and stockholder of Celgene and is a former employee and stockholder of GlaxoSmithKline. Dr. Shanmugasundaram is an employee and stockholder of Celgene and a stockholder of Pfizer. Celgene has now become a wholly-owned subsidiary of Bristol-Myers Squibb. Dr. Aschenbrenner reported grants from UCB Pharma GmbH and grants from Eli Lilly and Company outside the submitted work. Dr. Uhlig reported grants from Celgene during the conduct of the study, grants from UCB Pharma, grants from Eli Lilly, personal fees from AbbVie, personal fees from Pfizer, and "other" from Regeneron outside the submitted work. No other disclosures were reported.
Figures




References
-
- Dagoneau N., Scheffer D., Huber C., Al-Gazali L.I., Di Rocco M., Godard A., Martinovic J., Raas-Rothschild A., Sigaudy S., Unger S., et al. . 2004. Null leukemia inhibitory factor receptor (LIFR) mutations in Stuve-Wiedemann/Schwartz-Jampel type 2 syndrome. Am. J. Hum. Genet. 74:298–305. 10.1086/381715 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

