CD4+ T Follicular Helper Cells in Human Tonsils and Blood Are Clonally Convergent but Divergent from Non-Tfh CD4+ Cells
- PMID: 31914381
- PMCID: PMC7029615
- DOI: 10.1016/j.celrep.2019.12.016
CD4+ T Follicular Helper Cells in Human Tonsils and Blood Are Clonally Convergent but Divergent from Non-Tfh CD4+ Cells
Abstract
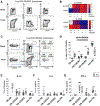
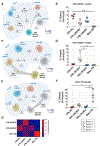
T follicular helper (Tfh) cells are fundamental for B cell selection and antibody maturation in germinal centers. Circulating Tfh (cTfh) cells constitute a minor proportion of the CD4+ T cells in peripheral blood, but their clonotypic relationship to Tfh populations resident in lymph nodes and the extent to which they differ from non-Tfh CD4+ cells have been unclear. Using donor-matched blood and tonsil samples, we investigate T cell receptor (TCR) sharing between tonsillar Tfh cells and peripheral Tfh and non-Tfh cell populations. TCR transcript sequencing reveals considerable clonal overlap between peripheral and tonsillar Tfh cell subsets as well as a clear distinction between Tfh and non-Tfh cells. Furthermore, influenza-specific cTfh cell clones derived from blood can be found in the repertoire of tonsillar Tfh cells. Therefore, human blood samples can be used to gain insight into the specificity of Tfh responses occurring in lymphoid tissues, provided that cTfh subsets are studied.
Keywords: T follicular helper cells; TCR repertoire; blood; influenza; tonsil.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
The authors declare no competing interests.
Figures





References
-
- Ansel KM, Ngo VN, Hyman PL, Luther SA, Förster R, Sedgwick JD, Browning JL, Lipp M, and Cyster JG (2000). A chemokine-driven positive feedback loop organizes lymphoid follicles. Nature 406, 309–314. - PubMed
-
- Avery DT, Bryant VL, Ma CS, de Waal Malefyt R, and Tangye SG (2008). IL-21-induced isotype switching to IgG and IgA by human naive B cells is differentially regulated by IL-4. J. Immunol 181, 1767–1779. - PubMed
-
- Baumjohann D, Preite S, Reboldi A, Ronchi F, Ansel KM, Lanzavecchia A, and Sallusto F (2013). Persistent antigen and germinal center B cells sustain T follicular helper cell responses and phenotype. Immunity 38, 596–605. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

