DNA sequence repeats identify numerous Type I restriction-modification systems that are potential epigenetic regulators controlling phase-variable regulons; phasevarions
- PMID: 31914596
- PMCID: PMC7383803
- DOI: 10.1096/fj.201901536RR
DNA sequence repeats identify numerous Type I restriction-modification systems that are potential epigenetic regulators controlling phase-variable regulons; phasevarions
Abstract
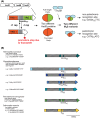
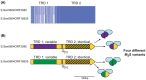
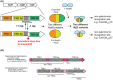
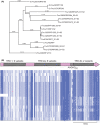
Over recent years several examples of randomly switching methyltransferases, associated with Type III restriction-modification (R-M) systems, have been described in pathogenic bacteria. In every case examined, changes in simple DNA sequence repeats result in variable methyltransferase expression and result in global changes in gene expression, and differentiation of the bacterial cell into distinct phenotypes. These epigenetic regulatory systems are called phasevarions, phase-variable regulons, and are widespread in bacteria, with 17.4% of Type III R-M system containing simple DNA sequence repeats. A distinct, recombination-driven random switching system has also been described in Streptococci in Type I R-M systems that also regulate gene expression. Here, we interrogate the most extensive and well-curated database of R-M systems, REBASE, by searching for all possible simple DNA sequence repeats in the hsdRMS genes that encode Type I R-M systems. We report that 7.9% of hsdS, 2% of hsdM, and of 4.3% of hsdR genes contain simple sequence repeats that are capable of mediating phase variation. Phase variation of both hsdM and hsdS genes will lead to differential methyltransferase expression or specificity, and thereby the potential to control phasevarions. These data suggest that in addition to well characterized phasevarions controlled by Type III mod genes, and the previously described Streptococcal Type I R-M systems that switch via recombination, approximately 10% of all Type I R-M systems surveyed herein have independently evolved the ability to randomly switch expression via simple DNA sequence repeats.
Keywords: R‐M systems; bacterial pathogenesis; epigenetics; phase variation; phasevarion.
© 2019 The Authors. The FASEB Journal published by Wiley Periodicals, Inc. on behalf of Federation of American Societies for Experimental Biology.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Systematic Analysis of REBASE Identifies Numerous Type I Restriction-Modification Systems with Duplicated, Distinct hsdS Specificity Genes That Can Switch System Specificity by Recombination.mSystems. 2020 Jul 28;5(4):e00497-20. doi: 10.1128/mSystems.00497-20. mSystems. 2020. PMID: 32723795 Free PMC article.
-
A survey of Type III restriction-modification systems reveals numerous, novel epigenetic regulators controlling phase-variable regulons; phasevarions.Nucleic Acids Res. 2018 Apr 20;46(7):3532-3542. doi: 10.1093/nar/gky192. Nucleic Acids Res. 2018. PMID: 29554328 Free PMC article.
-
Streptococcus suis Encodes Multiple Allelic Variants of a Phase-Variable Type III DNA Methyltransferase, ModS, That Control Distinct Phasevarions.mSphere. 2021 May 12;6(3):e00069-21. doi: 10.1128/mSphere.00069-21. mSphere. 2021. PMID: 33980672 Free PMC article.
-
Phasevarions of bacterial pathogens - phase-variable epigenetic regulators evolving from restriction-modification systems.Microbiology (Reading). 2019 Sep;165(9):917-928. doi: 10.1099/mic.0.000805. Epub 2019 Apr 17. Microbiology (Reading). 2019. PMID: 30994440 Review.
-
Epigenetic Regulation of Virulence and Immunoevasion by Phase-Variable Restriction-Modification Systems in Bacterial Pathogens.Annu Rev Microbiol. 2020 Sep 8;74:655-671. doi: 10.1146/annurev-micro-090817-062346. Epub 2020 Jul 20. Annu Rev Microbiol. 2020. PMID: 32689914 Review.
Cited by
-
Conserved DNA Methyltransferases: A Window into Fundamental Mechanisms of Epigenetic Regulation in Bacteria.Trends Microbiol. 2021 Jan;29(1):28-40. doi: 10.1016/j.tim.2020.04.007. Epub 2020 May 13. Trends Microbiol. 2021. PMID: 32417228 Free PMC article. Review.
-
The defensome of complex bacterial communities.Nat Commun. 2024 Mar 8;15(1):2146. doi: 10.1038/s41467-024-46489-0. Nat Commun. 2024. PMID: 38459056 Free PMC article.
-
Systematic Analysis of REBASE Identifies Numerous Type I Restriction-Modification Systems with Duplicated, Distinct hsdS Specificity Genes That Can Switch System Specificity by Recombination.mSystems. 2020 Jul 28;5(4):e00497-20. doi: 10.1128/mSystems.00497-20. mSystems. 2020. PMID: 32723795 Free PMC article.
-
Natural Recombination among Type I Restriction-Modification Systems Creates Diverse Genomic Methylation Patterns among Xylella fastidiosa Strains.Appl Environ Microbiol. 2023 Jan 31;89(1):e0187322. doi: 10.1128/aem.01873-22. Epub 2023 Jan 4. Appl Environ Microbiol. 2023. PMID: 36598481 Free PMC article.
-
Intragenic DNA inversions expand bacterial coding capacity.Nature. 2024 Oct;634(8032):234-242. doi: 10.1038/s41586-024-07970-4. Epub 2024 Sep 25. Nature. 2024. PMID: 39322669
References
-
- Moxon R, Bayliss C, Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Ann Rev Genet. 2006;40:307‐333. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

