Adeno-Associated Virus Serotype-Specific Inverted Terminal Repeat Sequence Role in Vector Transgene Expression
- PMID: 31914802
- PMCID: PMC7047122
- DOI: 10.1089/hum.2019.274
Adeno-Associated Virus Serotype-Specific Inverted Terminal Repeat Sequence Role in Vector Transgene Expression
Abstract
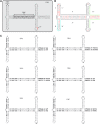
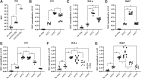
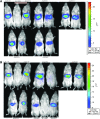
Adeno-associated viral vectors have been successfully used in laboratory and clinical settings for efficient gene delivery. In these vectors, 96% of the adeno-associated virus (AAV) genome is replaced with a gene cassette of interest, leaving only the 145 bp inverted terminal repeat (ITR) sequences. These cis-elements, primarily from AAV serotype 2, are required for genome rescue, replication, packaging, and vector persistence. Previous work from our lab and others have demonstrated that the AAV ITR2 sequence has inherent transcriptional activity, which may confound intended transgene expression in therapeutic applications. Currently, AAV capsids are extensively study for vector contribution; however, a comprehensive analysis of ITR promoter activity of various AAV serotypes has not been described to date. Here, the transcriptional activity of AAV ITRs from different serotypes (1-4, 6, and 7) was compared in numerous cell lines and a mouse model. Under the conditions used here, all ITRs tested were capable of promoting transgene expression both in vitro and in vivo. However, we observed three classes of AAV ITR expression in vitro. Class I ITRs (AAV2 and 3) generated the highest level, whereas class II (AAV 4) had intermediate levels, and class III (AAV1 and 6) had the lowest levels. These expression levels were consistent across multiple cell lines. Only ITR7 demonstrated cell-type dependent transcriptional activity. In vivo, all classes had promoter activity. Next-generation sequencing revealed multiple transcriptional start sites that originated from the ITR sequence, with most arising from within the Rep binding element. The collective results demonstrate that the serotype ITR sequence may have multiple levels of influence on transgene expression cassettes independent of promoter selection.
Keywords: AAV; ITR; promoter.
Conflict of interest statement
M.L.H. is an inventor on technology not assessed herein that has been licensed to Asklepios BioPharmaceutical and Tamid Bio. M.L.H. has also received royalties from Asklepios BioPharmaceutical related to a patent (9447433). M.L.H. and C.L. are co-founders of Bedrock Therapeutics. R.J.S. is the founder of and a shareholder at Asklepios BioPharmaceutical and Bamboo Therapeutics, Inc. He holds a patent (9475845) that has been licensed by the University of North Carolina at Chapel Hill to Asklepios BioPharmaceutical, for which he receives royalties. He has consulted for Baxter and has received payment for speaking. L.F.E. is currently an employee at Shape Therapeutics.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

