Global evaluation of taxonomic relationships and admixture within the Culex pipiens complex of mosquitoes
- PMID: 31915057
- PMCID: PMC6950815
- DOI: 10.1186/s13071-020-3879-8
Global evaluation of taxonomic relationships and admixture within the Culex pipiens complex of mosquitoes
Abstract
Background: Within the Culex pipiens mosquito complex, there are six contemporarily recognized taxa: Cx. quinquefasciatus, Cx. pipiens f. pipiens, Cx. pipiens f. molestus, Cx. pipiens pallens, Cx. australicus and Cx. globocoxitus. Many phylogenetic aspects within this complex have eluded resolution, such as the relationship of the two Australian endemic taxa to the other four members, as well as the evolutionary origins and taxonomic status of Cx. pipiens pallens and Cx. pipiens f. molestus. Ultimately, insights into lineage relationships within the complex will facilitate a better understanding of differential disease transmission by these mosquitoes. To this end, we have combined publicly available data with our own sequencing efforts to examine these questions.
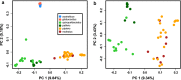
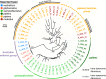
Results: We found that the two Australian endemic complex members, Cx. australicus and Cx. globocoxitus, comprise a monophyletic group, are genetically distinct, and are most closely related to the cosmopolitan Cx. quinquefasciatus. Our results also show that Cx. pipiens pallens is genetically distinct, but may have arisen from past hybridization. Lastly, we observed complicated patterns of genetic differentiation within and between Cx. pipiens f. pipiens and Cx. pipiens f. molestus.
Conclusions: Two Australian endemic Culex taxa, Cx. australicus and Cx. globocoxitus, belong within the Cx. pipiens complex, but have a relatively older evolutionary origin. They likely diverged from Cx. quinquefasciatus after its colonization of Australia. The taxon Cx. pipiens pallens is a distinct evolutionary entity that likely arose from past hybridization between Cx. quinquefasciatus and Cx. pipiens f. pipiens/Cx. pipiens f. molestus. Our results do not suggest it derives from ongoing hybridization. Finally, genetic differentiation within the Cx. pipiens f. pipiens and Cx. pipiens f. molestus samples suggests that they collectively form two separate geographic clades, one in North America and one in Europe and the Mediterranean. This may indicate that the Cx. pipiens f. molestus form has two distinct origins, arising from Cx. pipiens f. pipiens in each region. However, ongoing genetic exchange within and between these taxa have obscured their evolutionary histories, and could also explain the absence of monophyly among our samples. Overall, this work suggests many avenues that warrant further investigation.
Keywords: Culicidae; Disease vector; Genetic exchange; Mosquito; Population structure; Species complex.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
-
- Hey J, Waples RS, Arnold ML, Butlin RK, Harrison RG. Understanding and confronting species uncertainty in biology and conservation. Trends Ecol Evol. 2003;18:597–603. doi: 10.1016/j.tree.2003.08.014. - DOI
-
- Arnold ML. Divergence with genetic exchange. Oxford: Oxford University Press; 2015.
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

