Strains used in whole organism Plasmodium falciparum vaccine trials differ in genome structure, sequence, and immunogenic potential
- PMID: 31915075
- PMCID: PMC6950926
- DOI: 10.1186/s13073-019-0708-9
Strains used in whole organism Plasmodium falciparum vaccine trials differ in genome structure, sequence, and immunogenic potential
Abstract
Background: Plasmodium falciparum (Pf) whole-organism sporozoite vaccines have been shown to provide significant protection against controlled human malaria infection (CHMI) in clinical trials. Initial CHMI studies showed significantly higher durable protection against homologous than heterologous strains, suggesting the presence of strain-specific vaccine-induced protection. However, interpretation of these results and understanding of their relevance to vaccine efficacy have been hampered by the lack of knowledge on genetic differences between vaccine and CHMI strains, and how these strains are related to parasites in malaria endemic regions.
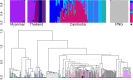
Methods: Whole genome sequencing using long-read (Pacific Biosciences) and short-read (Illumina) sequencing platforms was conducted to generate de novo genome assemblies for the vaccine strain, NF54, and for strains used in heterologous CHMI (7G8 from Brazil, NF166.C8 from Guinea, and NF135.C10 from Cambodia). The assemblies were used to characterize sequences in each strain relative to the reference 3D7 (a clone of NF54) genome. Strains were compared to each other and to a collection of clinical isolates (sequenced as part of this study or from public repositories) from South America, sub-Saharan Africa, and Southeast Asia.
Results: While few variants were detected between 3D7 and NF54, we identified tens of thousands of variants between NF54 and the three heterologous strains. These variants include SNPs, indels, and small structural variants that fall in regulatory and immunologically important regions, including transcription factors (such as PfAP2-L and PfAP2-G) and pre-erythrocytic antigens that may be key for sporozoite vaccine-induced protection. Additionally, these variants directly contributed to diversity in immunologically important regions of the genomes as detected through in silico CD8+ T cell epitope predictions. Of all heterologous strains, NF135.C10 had the highest number of unique predicted epitope sequences when compared to NF54. Comparison to global clinical isolates revealed that these four strains are representative of their geographic origin despite long-term culture adaptation; of note, NF135.C10 is from an admixed population, and not part of recently formed subpopulations resistant to artemisinin-based therapies present in the Greater Mekong Sub-region.
Conclusions: These results will assist in the interpretation of vaccine efficacy of whole-organism vaccines against homologous and heterologous CHMI.
Keywords: Genome assembly; Malaria; P. falciparum; PfSPZ vaccine; Whole-sporozoite vaccine.
Conflict of interest statement
T.L., B.K.L.S., and S.L.H. are salaried employees of Sanaria Inc., the developer and owner of PfSPZ vaccine, and the supplier of the PfSPZ strains sequenced in this study. In addition, S.L.H. and B.K.L.S. have a financial interest in Sanaria Inc. The remaining authors declare that they have no competing interests.
Figures





References
-
- World Malaria Report 2018. Geneva: World Health Organization; 2018 p. 1–210.
-
- Delemarre BJ, van der Kaay HJ. Tropical malaria contracted the natural way in the Netherlands. Ned Tijdschr Geneeskd. 1979;123(46):1981–1982. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 AI125579/AI/NIAID NIH HHS/United States
- R25 GM055036/GM/NIGMS NIH HHS/United States
- R44 AI058375/AI/NIAID NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- U19 AI110820/AI/NIAID NIH HHS/United States
- R44 AI055229/AI/NIAID NIH HHS/United States
- R01 AI141900/AI/NIAID NIH HHS/United States
- U19 AI089681/AI/NIAID NIH HHS/United States
- K01 HL140285/HL/NHLBI NIH HHS/United States
- U19 AI089683/AI/NIAID NIH HHS/United States
- ZIA HG200398/ImNIH/Intramural NIH HHS/United States
- 1ZIAHG200398/HG/NHGRI NIH HHS/United States
- F31 HL147471/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

