Dissemination of the blaNDM-5 Gene via IncX3-Type Plasmid among Enterobacteriaceae in Children
- PMID: 31915216
- PMCID: PMC6952193
- DOI: 10.1128/mSphere.00699-19
Dissemination of the blaNDM-5 Gene via IncX3-Type Plasmid among Enterobacteriaceae in Children
Abstract
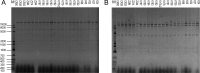
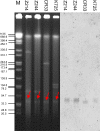
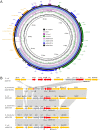
The continuous emergence of novel New Delhi metallo-β-lactamase-5 (NDM-5)-producing Enterobacteriaceae isolates is receiving more and more public attention. Twenty-two NDM-5-producing strains were identified from 146 carbapenemase-producing Enterobacteriaceae (CRE) strains isolated from pediatric patients between January and March 2017, indicating that the blaNDM-5 gene has spread to children. All 22 isolates, including 16 Klebsiella pneumoniae strains, four Klebsiella aerogenes strains, and two Escherichia coli strains, showed significantly high resistance to β-lactam antibiotics (except aztreonam) but remained susceptible to tigecycline and colistin. K. pneumoniae and K. aerogenes strains were respectively defined as homologous clonal isolates by pulsed-field gel electrophoresis (PFGE). Multilocus sequence typing (MLST) results confirmed the genetic relatedness with all K. pneumoniae strains belonging to sequence type (ST) 48. Two E. coli isolates (ST617 and ST1236) were considered genetically unrelated. Twenty-two blaNDM-5 plasmids were positive for the IncX3 amplicon and showed almost identical profiles after digestion with HindIII and EcoRI. Four representative strains (K. pneumoniae K725, K. aerogenes CR33, E. coli Z214, and E. coli Z244) were selected for further study. Plasmids harboring blaNDM-5 showed strong stability in both clinical isolates and transconjugants, without apparent plasmid loss after 100 serial generations. S1-PFGE followed by Southern blot analysis demonstrated that the blaNDM-5 gene was located on an ∼46-kb plasmid. Plasmid sequences of pNDM-K725, pNDM-CR33, and pNDM-Z214 were almost identical but were slightly different from that of pNDM-Z244. Compared with pNDM-Z244, ΔISAba125 and partial copies of IS3000 were missing. The genetic backgrounds of the blaNDM-5 gene in four strains were slightly different from that of the typical pNDM_MGR194. This study comprehensively characterized the horizontal gene transfer of the blaNDM-5 gene among different Enterobacteriaceae isolates in pediatric patients, and the IncX3-type plasmid was responsible for the spread.IMPORTANCE The emergence of CRE strains resistant to multiple antibiotics is considered a substantial threat to human health. Therefore, all the efforts to provide a detailed molecular transmission mechanism of specific drug resistance can contribute positively to prevent the further spread of multidrug-resistant bacteria. Although the new superbug harboring blaNDM-5 has been reported in many countries, it was mostly identified among E. coli strains, and the gene transfer mechanism has not been fully recognized and studied. In this work, we identified 22 blaNDM-5-positive strains in different species of Enterobacteriaceae, including 16 Klebsiella pneumoniae strains, four Klebsiella aerogenes strains, and two Escherichia coli strains, which indicated the horizontal gene transfer of blaNDM-5 among Enterobacteriaceae strains in pediatric patients. Moreover, blaNDM-5 was located on a 46-kb IncX3 plasmid, which is possibly responsible for this widespread horizontal gene transfer. The different genetic contexts of the blaNDM-5 gene indicated some minor evolutions of the plasmid, based on the complete sequences of the blaNDM-5 plasmids. These findings are of great significance to understand the transmission mechanism of drug resistance genes, develop anti-infection treatment, and take effective infection control measures.
Keywords: Enterobacterales; Enterobacteriaceae; IncX3-type plasmid; NDM-5; ST48; carbapenemase; children.
Copyright © 2020 Tian et al.
Figures

Similar articles
-
Dissemination and Stability of the blaNDM-5-Carrying IncX3-Type Plasmid among Multiclonal Klebsiella pneumoniae Isolates.mSphere. 2020 Nov 4;5(6):e00917-20. doi: 10.1128/mSphere.00917-20. mSphere. 2020. PMID: 33148824 Free PMC article.
-
Clone dissemination of IncX3 plasmid carrying blaNDM-1 in ST76 carbapenem resistance Klebsiella pneumoniae and bactericidal efficiency of aztreonam combined with avibactam in vitro and in vivo.J Glob Antimicrob Resist. 2024 Mar;36:244-251. doi: 10.1016/j.jgar.2023.12.031. Epub 2024 Jan 24. J Glob Antimicrob Resist. 2024. PMID: 38272211
-
Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China.Antimicrob Resist Infect Control. 2018 Apr 26;7:59. doi: 10.1186/s13756-018-0349-6. eCollection 2018. Antimicrob Resist Infect Control. 2018. PMID: 29713466 Free PMC article.
-
Systematic review of carbapenem-resistant Enterobacteriaceae causing neonatal sepsis in China.Ann Clin Microbiol Antimicrob. 2019 Nov 14;18(1):36. doi: 10.1186/s12941-019-0334-9. Ann Clin Microbiol Antimicrob. 2019. PMID: 31727088 Free PMC article.
-
New tools to mitigate drug resistance in Enterobacteriaceae - Escherichia coli and Klebsiella pneumoniae.Crit Rev Microbiol. 2023 Aug;49(4):435-454. doi: 10.1080/1040841X.2022.2080525. Epub 2022 Jun 1. Crit Rev Microbiol. 2023. PMID: 35649163 Review.
Cited by
-
Emergence of a Novel NDM-5-Producing Sequence Type 4523 Klebsiella pneumoniae Strain Causing Bloodstream Infection in China.Microbiol Spectr. 2022 Oct 26;10(5):e0084222. doi: 10.1128/spectrum.00842-22. Epub 2022 Aug 22. Microbiol Spectr. 2022. PMID: 35993711 Free PMC article.
-
Genomic evolution and dissemination of non-conjugative virulence plasmid of ST65 carbapenem-resistant and hypervirulent Klebsiella pneumoniae strains in a Chinese hospital.Front Cell Infect Microbiol. 2025 Jun 12;15:1548300. doi: 10.3389/fcimb.2025.1548300. eCollection 2025. Front Cell Infect Microbiol. 2025. PMID: 40575489 Free PMC article.
-
Exploring the Antibiotic Resistance Burden in Livestock, Livestock Handlers and Their Non-Livestock Handling Contacts: A One Health Perspective.Front Microbiol. 2021 Apr 20;12:651461. doi: 10.3389/fmicb.2021.651461. eCollection 2021. Front Microbiol. 2021. PMID: 33959112 Free PMC article.
-
In Vitro Activity of Cefiderocol on Multiresistant Bacterial Strains and Genomic Analysis of Two Cefiderocol Resistant Strains.Antibiotics (Basel). 2023 Apr 20;12(4):785. doi: 10.3390/antibiotics12040785. Antibiotics (Basel). 2023. PMID: 37107147 Free PMC article.
-
High prevalence of carbapenem-resistant Enterobacter cloacae complex in a tertiary hospital over a decade.Microbiol Spectr. 2024 Oct 30;12(12):e0078024. doi: 10.1128/spectrum.00780-24. Online ahead of print. Microbiol Spectr. 2024. PMID: 39475294 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
