A cell atlas of the adult Drosophila midgut
- PMID: 31915294
- PMCID: PMC6983450
- DOI: 10.1073/pnas.1916820117
A cell atlas of the adult Drosophila midgut
Abstract
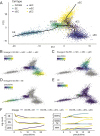
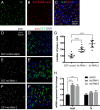
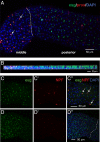
Studies of the adult Drosophila midgut have led to many insights in our understanding of cell-type diversity, stem cell regeneration, tissue homeostasis, and cell fate decision. Advances in single-cell RNA sequencing provide opportunities to identify new cell types and molecular features. We used single-cell RNA sequencing to characterize the transcriptome of midgut epithelial cells and identified 22 distinct clusters representing intestinal stem cells, enteroblasts, enteroendocrine cells (EEs), and enterocytes. This unbiased approach recovered most of the known intestinal stem cells/enteroblast and EE markers, highlighting the high quality of the dataset, and led to insights on intestinal stem cell biology, cell type-specific organelle features, the roles of new transcription factors in progenitors and regional variation along the gut, 5 additional EE gut hormones, EE hormonal expression diversity, and paracrine function of EEs. To facilitate mining of this rich dataset, we provide a web-based resource for visualization of gene expression in single cells. Altogether, our study provides a comprehensive resource for addressing functions of genes in the midgut epithelium.
Keywords: Drosophila; enteroblast; enteroendocrine cell; gut; stem cell.
Conflict of interest statement
The authors declare no competing interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

