Transcription factor NRF2 uses the Hippo pathway effector TAZ to induce tumorigenesis in glioblastomas
- PMID: 31918259
- PMCID: PMC7016245
- DOI: 10.1016/j.redox.2019.101425
Transcription factor NRF2 uses the Hippo pathway effector TAZ to induce tumorigenesis in glioblastomas
Abstract
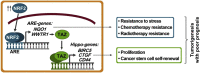
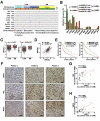
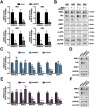
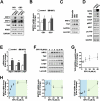
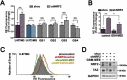
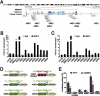
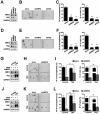
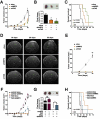
Transcription factor NRF2 orchestrates a cellular defense against oxidative stress and, so far, has been involved in tumor progression by providing a metabolic adaptation to tumorigenic demands and resistance to chemotherapeutics. In this study, we discover that NRF2 also propels tumorigenesis in gliomas and glioblastomas by inducing the expression of the transcriptional co-activator TAZ, a protein of the Hippo signaling pathway that promotes tumor growth. The expression of the genes encoding NRF2 (NFE2L2) and TAZ (WWTR1) showed a positive correlation in 721 gliomas from The Cancer Genome Atlas database. Moreover, NRF2 and TAZ protein levels also correlated in immunohistochemical tissue arrays of glioblastomas. Genetic knock-down of NRF2 decreased, while NRF2 overexpression or chemical activation with sulforaphane, increased TAZ transcript and protein levels. Mechanistically, we identified several NRF2-regulated functional enhancers in the regulatory region of WWTR1. The relevance of the new NRF2/TAZ axis in tumorigenesis was demonstrated in subcutaneous and intracranial grafts. Thus, intracranial inoculation of NRF2-depleted glioma stem cells did not develop tumors as determined by magnetic resonance imaging. Forced TAZ overexpression partly rescued both stem cell growth in neurospheres and tumorigenicity. Hence, NRF2 not only enables tumor cells to be competent to proliferate but it also propels tumorigenesis by activating the TAZ-mediated Hippo transcriptional program.
Keywords: Cancer stem cells; Chemotherapy; Glioblastoma; Oxidative stress.
Copyright © 2020 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare no competing interests.
Figures
References
-
- Cuadrado A., Manda G., Hassan A., Alcaraz M.J., Barbas C., Daiber A., Ghezzi P., Leon R., Lopez M.G., Oliva B., Pajares M., Rojo A.I., Robledinos-Anton N., Valverde A.M., Guney E., Schmidt H. Transcription factor NRF2 as a therapeutic target for chronic diseases: a systems medicine approach. Pharmacol. Rev. 2018;70:348–383. - PubMed
-
- Hayes J.D., Dinkova-Kostova A.T. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014;39:199–218. - PubMed
-
- Lee S.B., Sellers B.N., DeNicola G.M. The regulation of NRF2 by nutrient-responsive signaling and its role in anabolic cancer metabolism. Antioxidants Redox Signal. 2018;29:1774–1791. - PubMed
-
- DeNicola G.M., Karreth F.A., Humpton T.J., Gopinathan A., Wei C., Frese K., Mangal D., Yu K.H., Yeo C.J., Calhoun E.S., Scrimieri F., Winter J.M., Hruban R.H., Iacobuzio-Donahue C., Kern S.E., Blair I.A., Tuveson D.A. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475:106–109. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

