Sensing of HIV-1 by TLR8 activates human T cells and reverses latency
- PMID: 31919342
- PMCID: PMC6952430
- DOI: 10.1038/s41467-019-13837-4
Sensing of HIV-1 by TLR8 activates human T cells and reverses latency
Abstract
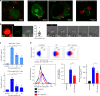
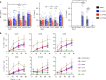
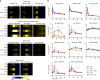
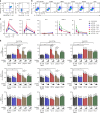
During HIV infection, cell-to-cell transmission results in endosomal uptake of the virus by target CD4+ T cells and potential exposure of the viral ssRNA genome to endosomal Toll-like receptors (TLRs). TLRs are instrumental in activating inflammatory responses in innate immune cells, but their function in adaptive immune cells is less well understood. Here we show that synthetic ligands of TLR8 boosted T cell receptor signaling, resulting in increased cytokine production and upregulation of surface activation markers. Adjuvant TLR8 stimulation, but not TLR7 or TLR9, further promoted T helper cell differentiation towards Th1 and Th17. In addition, we found that endosomal HIV induced cytokine secretion from CD4+ T cells in a TLR8-specific manner. TLR8 engagement also enhanced HIV-1 replication and potentiated the reversal of latency in patient-derived T cells. The adjuvant TLR8 activity in T cells can contribute to viral dissemination in the lymph node and low-grade inflammation in HIV patients. In addition, it can potentially be exploited for therapeutic targeting and vaccine development.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

