The nuclear pore complex prevents sister chromatid recombination during replicative senescence
- PMID: 31919430
- PMCID: PMC6952416
- DOI: 10.1038/s41467-019-13979-5
The nuclear pore complex prevents sister chromatid recombination during replicative senescence
Abstract
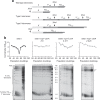
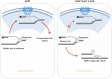
The Nuclear Pore Complex (NPC) has emerged as an important hub for processing various types of DNA damage. Here, we uncover that fusing a DNA binding domain to the NPC basket protein Nup1 reduces telomere relocalization to nuclear pores early after telomerase inactivation. This Nup1 modification also impairs the relocalization to the NPC of expanded CAG/CTG triplet repeats. Strikingly, telomerase negative cells bypass senescence when expressing this Nup1 modification by maintaining a minimal telomere length compatible with proliferation through rampant unequal exchanges between sister chromatids. We further report that a Nup1 mutant lacking 36 C-terminal residues recapitulates the phenotypes of the Nup1-LexA fusion indicating a direct role of Nup1 in the relocation of stalled forks to NPCs and restriction of error-prone recombination between repeated sequences. Our results reveal a new mode of telomere maintenance that could shed light on how 20% of cancer cells are maintained without telomerase or ALT.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

