Identification of tRNA-derived small RNA (tsRNA) responsive to the tumor suppressor, RUNX1, in breast cancer
- PMID: 31919859
- PMCID: PMC7238950
- DOI: 10.1002/jcp.29419
Identification of tRNA-derived small RNA (tsRNA) responsive to the tumor suppressor, RUNX1, in breast cancer
Abstract
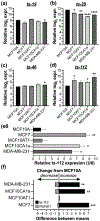
Despite recent advances in targeted therapies, the molecular mechanisms driving breast cancer initiation, progression, and metastasis are minimally understood. Growing evidence indicate that transfer RNA (tRNA)-derived small RNAs (tsRNA) contribute to biological control and aberrations associated with cancer development and progression. The runt-related transcription factor 1 (RUNX1) transcription factor is a tumor suppressor in the mammary epithelium whereas RUNX1 downregulation is functionally associated with breast cancer initiation and progression. We identified four tsRNA (ts-19, ts-29, ts-46, and ts-112) that are selectively responsive to expression of the RUNX1 tumor suppressor. Our finding that ts-112 and RUNX1 anticorrelate in normal-like mammary epithelial and breast cancer lines is consistent with tumor-related activity of ts-112 and tumor suppressor activity of RUNX1. Inhibition of ts-112 in MCF10CA1a aggressive breast cancer cells significantly reduced proliferation. Ectopic expression of a ts-112 mimic in normal-like mammary epithelial MCF10A cells significantly increased proliferation. These findings support an oncogenic potential for ts-112. Moreover, RUNX1 may repress ts-112 to prevent overactive proliferation in breast epithelial cells to augment its established roles in maintaining the mammary epithelium.
Keywords: RUNX1; breast cancer; ncRNA; tRNA-derived fragments (tRF); tRNA-derived small RNA (tsRNA).
© 2020 Wiley Periodicals, Inc.
Conflict of interest statement
Conflict of Interest Statement:
The authors declare no conflict of interests.
Figures
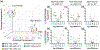


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

