Getting a handle on chemical probes of chomatin readers
- PMID: 31920100
- PMCID: PMC8025116
- DOI: 10.4155/fmc-2019-0274
Getting a handle on chemical probes of chomatin readers
Abstract
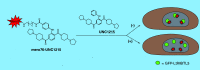
The dynamic nature of histone post-translational modifications such as methylation or acetylation makes possible the alteration of disease associated epigenetic states through the manipulation of the associated epigenetic machinery. One approach is through small molecule perturbation. Chemical probes of epigenetic reader domains have been critical in improving our understanding of the biological consequences of modulating their targets, while also enabling the development of novel probe-based reagents. By appending a functional handle to a reader domain probe, a chemical toolbox of reagents can be created to facilitate chemiprecipitation of epigenetic complexes, evaluate probe selectivity, develop in vitro screening assays, visualize cellular target localization, enable target degradation and recruit epigenetic machinery to a site within the genome in a highly controlled fashion.
Keywords: biotin; bivalent ligand; chemical probe; chemical tool; chromatin reader; epigenetics.
Conflict of interest statement
LI James gratefully acknowledges the National Institute on Drug Abuse, National Institutes of Health (NIH; grant R61DA047023), the National Cancer Institute, (NIH; grant R01CA242305), and the University Cancer Research Fund, University of North Carolina at Chapel Hill for support. JM Waybright acknowledges the National Cancer Institute for a training grant (grant T32CA217824). The authors have no other relevant affiliations of financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Figures




Similar articles
-
Chemical Tools with Fluorescence Switches for Verifying Epigenetic Modifications.Acc Chem Res. 2019 Oct 15;52(10):2849-2857. doi: 10.1021/acs.accounts.9b00349. Epub 2019 Oct 2. Acc Chem Res. 2019. PMID: 31577127 Review.
-
Selective targeting of epigenetic reader domains.Expert Opin Drug Discov. 2017 May;12(5):449-463. doi: 10.1080/17460441.2017.1303474. Epub 2017 Mar 14. Expert Opin Drug Discov. 2017. PMID: 28277835 Review.
-
Improved methods for targeting epigenetic reader domains of acetylated and methylated lysine.Curr Opin Chem Biol. 2021 Aug;63:132-144. doi: 10.1016/j.cbpa.2021.03.002. Epub 2021 Apr 11. Curr Opin Chem Biol. 2021. PMID: 33852996 Free PMC article. Review.
-
Studying Chromatin Epigenetics with Fluorescence Microscopy.Int J Mol Sci. 2022 Aug 12;23(16):8988. doi: 10.3390/ijms23168988. Int J Mol Sci. 2022. PMID: 36012253 Free PMC article. Review.
-
Epigenetic targets and drug discovery: part 1: histone methylation.Pharmacol Ther. 2014 Sep;143(3):275-94. doi: 10.1016/j.pharmthera.2014.03.007. Epub 2014 Apr 2. Pharmacol Ther. 2014. PMID: 24704322 Review.
Cited by
-
Cdyl Deficiency Brakes Neuronal Excitability and Nociception through Promoting Kcnb1 Transcription in Peripheral Sensory Neurons.Adv Sci (Weinh). 2022 Apr;9(10):e2104317. doi: 10.1002/advs.202104317. Epub 2022 Feb 4. Adv Sci (Weinh). 2022. PMID: 35119221 Free PMC article.
-
Trimethyllysine Reader Proteins Exhibit Widespread Charge-Agnostic Binding via Different Mechanisms to Cationic and Neutral Ligands.J Am Chem Soc. 2024 Feb 7;146(5):3086-3093. doi: 10.1021/jacs.3c10031. Epub 2024 Jan 24. J Am Chem Soc. 2024. PMID: 38266163 Free PMC article.
-
A Peptidomimetic Ligand Targeting the Chromodomain of MPP8 Reveals HRP2's Association with the HUSH Complex.ACS Chem Biol. 2021 Sep 17;16(9):1721-1736. doi: 10.1021/acschembio.1c00429. Epub 2021 Aug 20. ACS Chem Biol. 2021. PMID: 34415726 Free PMC article.
-
Negative Cooperativity in the UHRF1 TTD-PHD Dual Domain Masks the Contributions of Cation-π Interactions between Trimethyllysine and the TTD Aromatic Cage.Chemistry. 2025 May 19;31(28):e202500848. doi: 10.1002/chem.202500848. Epub 2025 Apr 27. Chemistry. 2025. PMID: 40202581
-
Targeting Epigenetic 'Readers' with Natural Compounds for Cancer Interception.J Cancer Prev. 2020 Dec 30;25(4):189-203. doi: 10.15430/JCP.2020.25.4.189. J Cancer Prev. 2020. PMID: 33409252 Free PMC article. Review.
References
-
- Garbaccio RM, Parmee ER. The impact of chemical probes in drug discovery: a pharmaceutical industry perspective. Cell Chem. Biol. 23(1), 10–17 (2016). - PubMed
-
- Frye SV. The art of the chemical probe. Nat. Chem. Biol. 6(3), 159–161 (2010). - PubMed
-
• Editorial explaining the criteria that defines a chemical probe.
-
- Schreiber SL. Small molecules: the missing link in the central dogma. Nat. Chem. Biol. 1(2), 64–66 (2005). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
