Quantitation and Comparison of Phenotypic Heterogeneity Among Single Cells of Monoclonal Microbial Populations
- PMID: 31921014
- PMCID: PMC6933826
- DOI: 10.3389/fmicb.2019.02814
Quantitation and Comparison of Phenotypic Heterogeneity Among Single Cells of Monoclonal Microbial Populations
Abstract
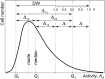
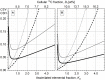
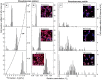
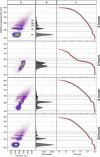
Phenotypic heterogeneity within microbial populations arises even when the cells are exposed to putatively constant and homogeneous conditions. The outcome of this phenomenon can affect the whole function of the population, resulting in, for example, new "adapted" metabolic strategies and impacting its fitness at given environmental conditions. Accounting for phenotypic heterogeneity becomes thus necessary, due to its relevance in medical and applied microbiology as well as in environmental processes. Still, a comprehensive evaluation of this phenomenon requires a common and unique method of quantitation, which allows for the comparison between different studies carried out with different approaches. Consequently, in this study, two widely applicable indices for quantitation of heterogeneity were developed. The heterogeneity coefficient (HC) is valid when the population follows unimodal activity, while the differentiation tendency index (DTI) accounts for heterogeneity implying outbreak of subpopulations and multimodal activity. We demonstrated the applicability of HC and DTI for heterogeneity quantitation on stable isotope probing with nanoscale secondary ion mass spectrometry (SIP-nanoSIMS), flow cytometry, and optical microscopy datasets. The HC was found to provide a more accurate and precise measure of heterogeneity, being at the same time consistent with the coefficient of variation (CV) applied so far. The DTI is able to describe the differentiation in single-cell activity within monoclonal populations resolving subpopulations with low cell abundance, individual cells with similar phenotypic features (e.g., isotopic content close to natural abundance, as detected with nanoSIMS). The developed quantitation approach allows for a better understanding on the impact and the implications of phenotypic heterogeneity in environmental, medical and applied microbiology, microbial ecology, cell biology, and biotechnology.
Keywords: SIP–nanoSIMS; Zipf's law; anabolic activity; flow cytometry; heterogeneity quantitation; multimodality; phenotypic heterogeneity; single-cell resolution.
Copyright © 2019 Calabrese, Voloshynovska, Musat, Thullner, Schlömann, Richnow, Lambrecht, Müller, Wick, Musat and Stryhanyuk.
Figures





References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

