Evidence of a Cellulosic Layer in Pandoravirus massiliensis Tegument and the Mystery of the Genetic Support of Its Biosynthesis
- PMID: 31921087
- PMCID: PMC6932959
- DOI: 10.3389/fmicb.2019.02932
Evidence of a Cellulosic Layer in Pandoravirus massiliensis Tegument and the Mystery of the Genetic Support of Its Biosynthesis
Abstract
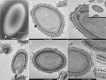
Pandoraviruses are giant viruses of ameba with 1 μm-long virions. They have an ovoid morphology and are surrounded by a tegument-like structure lacking any capsid protein nor any gene encoding a capsid protein. In this work, we studied the ultrastructure of the tegument surrounding Pandoravirus massiliensis virions and noticed that this tegument is composed of a peripheral sugar layer, an electron-dense membrane, and a thick electron-dense layer consisting in several tubules arranged in a helicoidal structure resembling that of cellulose. Pandoravirus massiliensis particles were stained by Calcofluor white, a fluorescent dye of cellulose, and the enzymatic treatment of particles by cellulase showed the degradation of the viral tegument. We first hypothesized that the cellulose tegument could be synthesized by enzymes encoded by the virus. Bioinformatic analyses revealed in P. massiliensis, a candidate gene encoding a putative cellulose synthase, with a homology with the BcsA domain, one of the catalytic subunits of the bacterial cellulose synthase, but with a low level of homology. This gene was transcribed during the replicative cycle of P. massiliensis, but several arguments run counter to this hypothesis. Indeed, even if this gene is present in other pandoraviruses, the one of the strain studied is the only one to have this BcsA domain and no other enzymes involved in the synthesis of cellulose could be detected, although we cannot rule out that such genes could have been undetected among the large proportion of Orfans of pandoraviruses. As an alternative, we investigated whether P. massiliensis could divert the cellulose synthesis machinery of the ameba to its own account. Indeed, contrary to what is observed in the case of infections with other giant viruses such as mimiviruses, it appears that the transcription of the ameba, at least for the cellulose synthase gene, continues throughout the growth phase of particles of P. massiliensis. Finally, we believe that this scenario is more plausible. If confirmed, it could be a unique mechanism in the virosphere.
Keywords: Pandoravirus; capsid; cellulose; cellulose synthase; giant virus.
Copyright © 2019 Brahim Belhaouari, Baudoin, Gnankou, Di Pinto, Colson, Aherfi and La Scola.
Figures








References
LinkOut - more resources
Full Text Sources
Research Materials

