A Snapshot of the Trehalose Pathway During Seed Imbibition in Medicago truncatula Reveals Temporal- and Stress-Dependent Shifts in Gene Expression Patterns Associated With Metabolite Changes
- PMID: 31921241
- PMCID: PMC6930686
- DOI: 10.3389/fpls.2019.01590
A Snapshot of the Trehalose Pathway During Seed Imbibition in Medicago truncatula Reveals Temporal- and Stress-Dependent Shifts in Gene Expression Patterns Associated With Metabolite Changes
Abstract
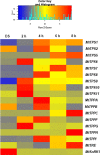
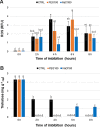
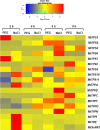
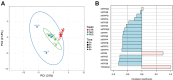
Trehalose, a non-reducing disaccharide with multiple functions, among which source of energy and carbon, stress protectant, and signaling molecule, has been mainly studied in relation to plant development and response to stress. The trehalose pathway is conserved among different organisms and is composed of three enzymes: trehalose-6-phosphate synthase (TPS), which converts uridine diphosphate (UDP)-glucose and glucose-6-phosphate to trehalose-6-phosphate (T6P), trehalose-6-phosphatase (TPP), which dephosphorylates T6P to produce trehalose, and trehalase (TRE), responsible for trehalose catabolism. In plants, the trehalose pathway has been mostly studied in resurrection plants and the model plant Arabidopsis thaliana, where 11 AtTPS, 10 AtTPP, and 1 AtTRE genes are present. Here, we aim to investigate the involvement of the trehalose pathway in the early stages of seed germination (specifically, seed imbibition) using the model legume Medicago truncatula as a working system. Since not all the genes belonging to the trehalose pathway had been identified in M. truncatula, we first conducted an in silico analysis using the orthologous gene sequences from A. thaliana. Nine MtTPSs, eight MtTPPs, and a single MtTRE gene were hereby identified. Subsequently, the expression profiles of all the genes (together with the sucrose master-regulator SnRK1) were investigated during seed imbibition with water or stress agents (polyethylene glycol and sodium chloride). The reported data show a temporal distribution and preferential expression of specific TPS and TPP isoforms during seed imbibition with water. Moreover, it was possible to distinguish a small set of genes (e.g., MtTPS1, MtTPS7, MtTPS10, MtTPPA, MtTPPI, MtTRE) having a potential impact as precocious hallmarks of the seed response to stress. When the trehalose levels were measured by high-performance liquid chromatography, a significant decrease was observed during seed imbibition, suggesting that trehalose may act as an energy source rather than osmoprotectant. This is the first report investigating the expression profiles of genes belonging to the trehalose pathway during seed imbibition, thus ascertaining their involvement in the pre-germinative metabolism and their potential as tools to improve seed germination efficiency.
Keywords: Medicago truncatula; abiotic stress; gene expression; seed imbibition; trehalose.
Copyright © 2019 Macovei, Pagano, Cappuccio, Gallotti, Dondi, De Sousa Araujo, Fevereiro and Balestrazzi.
Figures


References
LinkOut - more resources
Full Text Sources

