Alternative Splicing of Circadian Clock Genes Correlates With Temperature in Field-Grown Sugarcane
- PMID: 31921258
- PMCID: PMC6936171
- DOI: 10.3389/fpls.2019.01614
Alternative Splicing of Circadian Clock Genes Correlates With Temperature in Field-Grown Sugarcane
Abstract
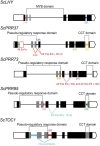
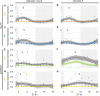
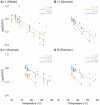
Alternative Splicing (AS) is a mechanism that generates different mature transcripts from precursor mRNAs (pre-mRNAs) of the same gene. In plants, a wide range of physiological and metabolic events are related to AS, as well as fast responses to changes in temperature. AS is present in around 60% of intron-containing genes in Arabidopsis, 46% in rice, and 38% in maize and it is widespread among the circadian clock genes. Little is known about how AS influences the circadian clock of C4 plants, like commercial sugarcane, a C4 crop with a complex hybrid genome. This work aims to test if the daily dynamics of AS forms of circadian clock genes are regulated by environmental factors, such as temperature, in the field. A systematic search for AS in five sugarcane clock genes, ScLHY, ScPRR37, ScPRR73, ScPRR95, and ScTOC1 using different organs of sugarcane sampled during winter, with 4 months old plants, and during summer, with 9 months old plants, revealed temperature- and organ-dependent expression of at least one alternatively spliced isoform in all genes. Expression of AS isoforms varied according to the season. Our results suggest that AS events in circadian clock genes are correlated with temperature.
Keywords: alternative splicing; circadian clock; diel rhythms; field experiment; gene expression; sugarcane.
Copyright © 2019 Dantas, Calixto, Dourado, Carneiro, Brown and Hotta.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

