Long Non-coding RNA LINC00114 Facilitates Colorectal Cancer Development Through EZH2/DNMT1-Induced miR-133b Suppression
- PMID: 31921641
- PMCID: PMC6928983
- DOI: 10.3389/fonc.2019.01383
Long Non-coding RNA LINC00114 Facilitates Colorectal Cancer Development Through EZH2/DNMT1-Induced miR-133b Suppression
Abstract
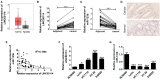
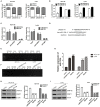
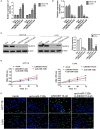
This study aimed to identify the roles of the long non-coding RNA LINC00114 in colorectal cancer (CRC) development. The expression levels of LINC00114 and miR-133b in CRC were determined by reverse transcription (RT)-polymerase chain reaction (PCR) and the functions of LINC00114 in CRC were evaluated in vitro and in vivo. Methylation-specific PCR assay was performed to detect the miR-133b promoter methylation in CRC cells. Bioinformatics analysis, RNA immunoprecipitation, dual luciferase assay, RNA pull-down, co-immunoprecipitation (IP), and chromatin IP (ChIP) assays were used to elucidate whether LINC00114 could recruit EZH2/DNMT1 and bind to the miR-133b promoter region, leading to dysregulated methylation and the depression of miR-133b. The expression levels of DNA methyltransferases (DNMTs), EZH2, and nucleoporin 214(NUP214) were analyzed by western blotting. Data showed that LINC00114 was highly expressed, whereas miR-133b was downregulated in the CRC tissues and cells. In vitro, silencing LINC00114 inhibited cell proliferation and impeded cell cycle at the G1/S phase by upregulating miR-133b. In vivo, LINC00114 knockdown reduced tumor growth. Further analysis showed that the methylation in miR-133b promoter region was increased in the CRC and silencing LINC00114 increased miR-133b expression through depressing methylation of its promoter region. ChIP-PCR experiments demonstrated that EZH2 and DNMT1 could bind to the miR-133b promoter region and it was abolished by LINC00114 knockdown. sh-EZH2 reversed the overexpression of DNMTs and CRC cell cycle progression induced by the LINC00114 upregulation. LINC00114 could regulate the NUP214 protein expression by sponging miR-133b. These results demonstrated that LINC00114 suppressed miR-133b expression via EZH2/DNMT1-mediated methylation of its promoter region, indicating that LINC00114 might be a potential novel target for CRC diagnosis and treatment.
Keywords: DNMT1; EZH2; LINC00114; colorectal cancer; methylation; miR-133b.
Copyright © 2019 Lv, He, Chen, Yu, Che, Tao, Wang, Jian and Zhang.
Figures




References
LinkOut - more resources
Full Text Sources

