Protein Amphipathic Helix Insertion: A Mechanism to Induce Membrane Fission
- PMID: 31921835
- PMCID: PMC6914677
- DOI: 10.3389/fcell.2019.00291
Protein Amphipathic Helix Insertion: A Mechanism to Induce Membrane Fission
Abstract
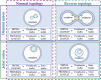
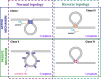
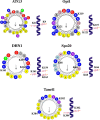
One of the fundamental features of biomembranes is the ability to fuse or to separate. These processes called respectively membrane fusion and fission are central in the homeostasis of events such as those related to intracellular membrane traffic. Proteins that contain amphipathic helices (AHs) were suggested to mediate membrane fission via shallow insertion of these helices into the lipid bilayer. Here we analyze the AH-containing proteins that have been identified as essential for membrane fission and categorize them in few subfamilies, including small GTPases, Atg proteins, and proteins containing either the ENTH/ANTH- or the BAR-domain. AH-containing fission-inducing proteins may require cofactors such as additional proteins (e.g., lipid-modifying enzymes), or lipids (e.g., phosphatidylinositol 4,5-bisphosphate [PtdIns(4,5)P2], phosphatidic acid [PA], or cardiolipin). Both PA and cardiolipin possess a cone shape and a negative charge (-2) that favor the recruitment of the AHs of fission-inducing proteins. Instead, PtdIns(4,5)P2 is characterized by an high negative charge able to recruit basic residues of the AHs of fission-inducing proteins. Here we propose that the AHs of fission-inducing proteins contain sequence motifs that bind lipid cofactors; accordingly (K/R/H)(K/R/H)xx(K/R/H) is a PtdIns(4,5)P2-binding motif, (K/R)x6(F/Y) is a cardiolipin-binding motif, whereas KxK is a PA-binding motif. Following our analysis, we show that the AHs of many fission-inducing proteins possess five properties: (a) at least three basic residues on the hydrophilic side, (b) ability to oligomerize, (c) optimal (shallow) depth of insertion into the membrane, (d) positive cooperativity in membrane curvature generation, and (e) specific interaction with one of the lipids mentioned above. These lipid cofactors favor correct conformation, oligomeric state and optimal insertion depth. The most abundant lipid in a given organelle possessing high negative charge (more negative than -1) is usually the lipid cofactor in the fission event. Interestingly, naturally occurring mutations have been reported in AH-containing fission-inducing proteins and related to diseases such as centronuclear myopathy (amphiphysin 2), Charcot-Marie-Tooth disease (GDAP1), Parkinson's disease (α-synuclein). These findings add to the interest of the membrane fission process whose complete understanding will be instrumental for the elucidation of the pathogenesis of diseases involving mutations in the protein AHs.
Keywords: amphipathic helix; fission-inducing protein; lipid cofactor; lipid-binding site; membrane fission; membrane scission; neck-hemifission model; shallow insertion.
Copyright © 2019 Zhukovsky, Filograna, Luini, Corda and Valente.
Figures



References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

