Alternative splicing programming of axon formation
- PMID: 31922356
- PMCID: PMC7594648
- DOI: 10.1002/wrna.1585
Alternative splicing programming of axon formation
Abstract
Alternative pre-mRNA splicing generates multiple mRNA isoforms of different structures and functions from a single gene. While the prevalence of alternative splicing control is widely recognized, and the underlying regulatory mechanisms have long been studied, the physiological relevance and biological necessity for alternative splicing are only slowly being revealed. Significant inroads have been made in the brain, where alternative splicing regulation is particularly pervasive and conserved. Various aspects of brain development and function (from neurogenesis, neuronal migration, synaptogenesis, to the homeostasis of neuronal activity) involve alternative splicing regulation. Recent studies have begun to interrogate the possible role of alternative splicing in axon formation, a neuron-exclusive morphological and functional characteristic. We discuss how alternative splicing plays an instructive role in each step of axon formation. Converging genetic, molecular, and cellular evidence from studies of multiple alternative splicing regulators in different systems shows that a biological process as complicated and unique as axon formation requires highly coordinated and specific alternative splicing events. This article is categorized under: RNA Processing > Splicing Regulation/Alternative Splicing RNA in Disease and Development > RNA in Development.
Keywords: Axon guidance; CELF; NOVA; PTBP; RBFOX.
© 2020 Wiley Periodicals, Inc.
Conflict of interest statement
No conflict of interest
Figures
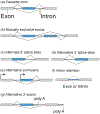




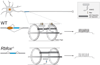
References
-
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, … Blencowe BJ (2012). The evolutionary landscape of alternative splicing in vertebrate species. Science, 338(6114), 1587–1593. - PubMed
-
- Black DL (2003). Mechanisms of alternative pre-messenger RNA splicing. Annual Review of Biochemistry, 72, 291–336. - PubMed
-
- Blencowe BJ (2017). [Review of The Relationship between Alternative Splicing and Proteomic Complexity]. Trends in biochemical sciences, 42(6), 407–408. - PubMed
-
- Bray NL, Pimentel H, Melsted P, & Pachter L (2016). Near-optimal probabilistic RNA-seq quantification. Nature Biotechnology, 34(5), 525–527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

