Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome
- PMID: 31922488
- PMCID: PMC7039682
- DOI: 10.7554/eLife.53199
Structural basis for COMPASS recognition of an H2B-ubiquitinated nucleosome
Abstract
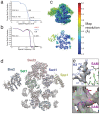
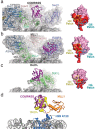
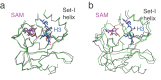
Methylation of histone H3K4 is a hallmark of actively transcribed genes that depends on mono-ubiquitination of histone H2B (H2B-Ub). H3K4 methylation in yeast is catalyzed by Set1, the methyltransferase subunit of COMPASS. We report here the cryo-EM structure of a six-protein core COMPASS subcomplex, which can methylate H3K4 and be stimulated by H2B-Ub, bound to a ubiquitinated nucleosome. Our structure shows that COMPASS spans the face of the nucleosome, recognizing ubiquitin on one face of the nucleosome and methylating H3 on the opposing face. As compared to the structure of the isolated core complex, Set1 undergoes multiple structural rearrangements to cement interactions with the nucleosome and with ubiquitin. The critical Set1 RxxxRR motif adopts a helix that mediates bridging contacts between the nucleosome, ubiquitin and COMPASS. The structure provides a framework for understanding mechanisms of trans-histone cross-talk and the dynamic role of H2B ubiquitination in stimulating histone methylation.
Keywords: S. cerevisiae; chromatin; chromosomes; gene expression; histone methylation; molecular biophysics; structural biology; transcription; ubiquitin.
© 2020, Worden et al.
Conflict of interest statement
EW, XZ No competing interests declared, CW Senior editor, eLife
Figures









Comment in
-
Navigating the structure of COMPASS.Elife. 2020 Feb 24;9:e54767. doi: 10.7554/eLife.54767. Elife. 2020. PMID: 32091392 Free PMC article.
Similar articles
-
Structural Basis of H2B Ubiquitination-Dependent H3K4 Methylation by COMPASS.Mol Cell. 2019 Dec 5;76(5):712-723.e4. doi: 10.1016/j.molcel.2019.10.013. Epub 2019 Nov 13. Mol Cell. 2019. PMID: 31733991 Free PMC article.
-
Histone H2B C-terminal helix mediates trans-histone H3K4 methylation independent of H2B ubiquitination.Mol Cell Biol. 2010 Jul;30(13):3216-32. doi: 10.1128/MCB.01008-09. Epub 2010 May 3. Mol Cell Biol. 2010. PMID: 20439497 Free PMC article.
-
Structural basis of recognition and destabilization of the histone H2B ubiquitinated nucleosome by the DOT1L histone H3 Lys79 methyltransferase.Genes Dev. 2019 Jun 1;33(11-12):620-625. doi: 10.1101/gad.323790.118. Epub 2019 Mar 28. Genes Dev. 2019. PMID: 30923167 Free PMC article.
-
The Core Complex of Yeast COMPASS and Human Mixed-Lineage Leukemia (MLL), Structure, Function, and Recognition of the Nucleosome.Subcell Biochem. 2024;104:101-117. doi: 10.1007/978-3-031-58843-3_6. Subcell Biochem. 2024. PMID: 38963485 Review.
-
Histone H2B ubiquitination and beyond: Regulation of nucleosome stability, chromatin dynamics and the trans-histone H3 methylation.Epigenetics. 2010 Aug 16;5(6):460-8. doi: 10.4161/epi.5.6.12314. Epub 2010 Aug 16. Epigenetics. 2010. PMID: 20523115 Free PMC article. Review.
Cited by
-
Mechanism for DPY30 and ASH2L intrinsically disordered regions to modulate the MLL/SET1 activity on chromatin.Nat Commun. 2021 May 19;12(1):2953. doi: 10.1038/s41467-021-23268-9. Nat Commun. 2021. PMID: 34012049 Free PMC article.
-
Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.Cell Discov. 2023 Dec 5;9(1):120. doi: 10.1038/s41421-023-00620-5. Cell Discov. 2023. PMID: 38052811 Free PMC article.
-
Distinct requirements for the COMPASS core subunits Set1, Swd1, and Swd3 during meiosis in the budding yeast Saccharomyces cerevisiae.G3 (Bethesda). 2021 Oct 19;11(11):jkab283. doi: 10.1093/g3journal/jkab283. G3 (Bethesda). 2021. PMID: 34849786 Free PMC article.
-
Loss of histone methyltransferase SETD1B in oogenesis results in the redistribution of genomic histone 3 lysine 4 trimethylation.Nucleic Acids Res. 2022 Feb 28;50(4):1993-2004. doi: 10.1093/nar/gkac051. Nucleic Acids Res. 2022. PMID: 35137160 Free PMC article.
-
Chromatin Regulation through Ubiquitin and Ubiquitin-like Histone Modifications.Trends Biochem Sci. 2021 Apr;46(4):258-269. doi: 10.1016/j.tibs.2020.11.005. Epub 2020 Dec 9. Trends Biochem Sci. 2021. PMID: 33308996 Free PMC article. Review.
References
-
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX : a comprehensive Python-based system for macromolecular structure solution. Acta Crystallographica Section D Biological Crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

