Cascading After Peridiagnostic Cancer Genetic Testing: An Alternative to Population-Based Screening
- PMID: 31922925
- PMCID: PMC7193752
- DOI: 10.1200/JCO.19.02010
Cascading After Peridiagnostic Cancer Genetic Testing: An Alternative to Population-Based Screening
Abstract
Purpose: Despite advances in DNA sequencing technology and expanded medical guidelines, the vast majority of individuals carrying pathogenic variants of common cancer susceptibility genes have yet to be identified. An alternative to population-wide genetic screening of healthy individuals would exploit the trend for genetic testing at the time of cancer diagnosis to guide therapy and prevention, combined with augmented familial diffusion or "cascade" of genomic risk information.
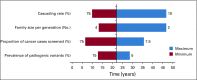
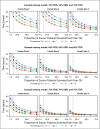
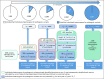
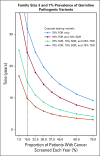
Methods: Using a multiple linear regression model, we derived the time interval to detect an estimated 3.9 million individuals in the United States with a pathogenic variant in 1 of 18 cancer susceptibility genes. We analyzed the impact of the proportion of incident patients sequenced, varying observed frequencies of pathogenic germline variants in patients with cancer, differential rates of diffusion of genetic information in families, and family size.
Results: The time to detect inherited cancer predisposing variants in the population is affected by the extent of cascade to first-, second-, and third-degree relatives (FDR, SDR, TDR, respectively), family size, prevalence of mutations in patients with cancer, and the proportion of patients with cancer sequenced. In a representative scenario, assuming a 7% prevalence of pathogenic variants across cancer types, an average family size of 3 per generation, and 15% of incident patients with cancer in the United States undergoing germline testing, the time to detect all 3.9 million individuals with pathogenic variants in 18 cancer susceptibility genes would be 46.2, 22.3, 13.6, and 9.9 years if 10%, 25%, 50%, and 70%, respectively, of all FDR, SDR, and TDR were tested for familial mutations.
Conclusion: Peridiagnostic and cascade cancer genetic testing offers an alternative strategy to achieve population-wide identification of cancer susceptibility mutations.
Figures
Comment in
-
Emerging Opportunity of Cascade Genetic Testing for Population-Wide Cancer Prevention and Control.J Clin Oncol. 2020 May 1;38(13):1371-1374. doi: 10.1200/JCO.20.00140. Epub 2020 Feb 25. J Clin Oncol. 2020. PMID: 32097078 No abstract available.
-
Peridiagnostic and cascade cancer genetic testing.Nat Rev Clin Oncol. 2020 May;17(5):277-278. doi: 10.1038/s41571-020-0348-4. Nat Rev Clin Oncol. 2020. PMID: 32152486 No abstract available.
-
Costs and Barriers of Cancer Screening After Positive Genetic Testing: Are Actionable Mutations Becoming "Unactionable"?J Natl Compr Canc Netw. 2022 Jun;20(6):616-617. doi: 10.6004/jnccn.2022.7013. J Natl Compr Canc Netw. 2022. PMID: 35714670 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

