Non-gradient and genotype-dependent patterns of RSV gene expression
- PMID: 31923213
- PMCID: PMC6953876
- DOI: 10.1371/journal.pone.0227558
Non-gradient and genotype-dependent patterns of RSV gene expression
Abstract
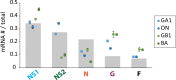
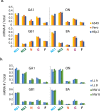
Respiratory syncytial virus (RSV) is a nonsegmented negative-strand RNA virus (NSV) and a leading cause of severe lower respiratory tract illness in infants and the elderly. Transcription of the ten RSV genes proceeds sequentially from the 3' promoter and requires conserved gene start (GS) and gene end (GE) signals. Previous studies using the prototypical GA1 genotype Long and A2 strains have indicated a gradient of gene transcription extending across the genome, with the highest level of mRNA coming from the most promoter-proximal gene, the first nonstructural (NS1) gene, and mRNA levels from subsequent genes dropping until reaching a minimum at the most promoter-distal gene, the polymerase (L) gene. However, recent reports show non-gradient levels of mRNA, with higher than expected levels from the attachment (G) gene. It is unknown to what extent different transcript stabilities might shape measured mRNA levels. It is also unclear whether patterns of RSV gene expression vary, or show strain- or genotype-dependence. To address this, mRNA abundances from five RSV genes were measured by quantitative real-time PCR (qPCR) in three cell lines and in cotton rats infected with RSV isolates belonging to four genotypes (GA1, ON, GB1, BA). Relative mRNA levels reached steady-state between four and 24 hours post-infection. Steady-state patterns were non-gradient and genotype-specific, where mRNA levels from the G gene exceeded those from the more promoter-proximal nucleocapsid (N) gene across isolates. Transcript stabilities could not account for the non-gradient patterns observed, indicating that relative mRNA levels more strongly reflect transcription than decay. Our results indicate that gene expression from a small but diverse set of RSV genotypes is non-gradient and genotype-dependent. We propose novel models of RSV transcription that can account for non-gradient transcription.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Effects of altering the transcription termination signals of respiratory syncytial virus on viral gene expression and growth in vitro and in vivo.J Virol. 2004 Jan;78(2):692-9. doi: 10.1128/jvi.78.2.692-699.2004. J Virol. 2004. PMID: 14694100 Free PMC article.
-
A Respiratory Syncytial Virus Attachment Gene Variant Associated with More Severe Disease in Infants Decreases Fusion Protein Expression, Which May Facilitate Immune Evasion.J Virol. 2020 Dec 22;95(2):e01201-20. doi: 10.1128/JVI.01201-20. Print 2020 Dec 22. J Virol. 2020. PMID: 33115881 Free PMC article.
-
Role of the M2-1 transcription antitermination protein of respiratory syncytial virus in sequential transcription.J Virol. 1999 Jul;73(7):5852-64. doi: 10.1128/JVI.73.7.5852-5864.1999. J Virol. 1999. PMID: 10364337 Free PMC article.
-
Structural Insights into the Respiratory Syncytial Virus RNA Synthesis Complexes.Viruses. 2021 May 5;13(5):834. doi: 10.3390/v13050834. Viruses. 2021. PMID: 34063087 Free PMC article. Review.
-
Respiratory Syncytial Virus: The Influence of Serotype and Genotype Variability on Clinical Course of Infection.Int J Mol Sci. 2017 Aug 6;18(8):1717. doi: 10.3390/ijms18081717. Int J Mol Sci. 2017. PMID: 28783078 Free PMC article. Review.
Cited by
-
Cryo-Electron Microscopy Structures of the Pneumoviridae Polymerases.Viral Immunol. 2021 Jan-Feb;34(1):18-26. doi: 10.1089/vim.2020.0018. Epub 2020 May 19. Viral Immunol. 2021. PMID: 32429800 Free PMC article.
-
Respiratory syncytial virus (RSV) enhances translation of virus-resembling AU-rich host transcripts.Virol J. 2025 Jul 15;22(1):244. doi: 10.1186/s12985-025-02838-z. Virol J. 2025. PMID: 40665364 Free PMC article.
-
Examining intra-host genetic variation of RSV by short read high-throughput sequencing.bioRxiv [Preprint]. 2024 Sep 3:2023.05.17.541198. doi: 10.1101/2023.05.17.541198. bioRxiv. 2024. PMID: 39282457 Free PMC article. Preprint.
-
Complete Genomic Characterization of Global Pathogens, Respiratory Syncytial Virus (RSV), and Human Norovirus (HuNoV) Using Probe-based Capture Enrichment.bioRxiv [Preprint]. 2024 Sep 16:2024.09.16.613242. doi: 10.1101/2024.09.16.613242. bioRxiv. 2024. Update in: Sci Rep. 2025 Jul 1;15(1):20526. doi: 10.1038/s41598-025-03398-6. PMID: 39345650 Free PMC article. Updated. Preprint.
-
Respiratory syncytial virus M2-1 protein associates non-specifically with viral messenger RNA and with specific cellular messenger RNA transcripts.PLoS Pathog. 2021 May 18;17(5):e1009589. doi: 10.1371/journal.ppat.1009589. eCollection 2021 May. PLoS Pathog. 2021. PMID: 34003848 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

