Diversity and plant growth-promoting functions of diazotrophic/N-scavenging bacteria isolated from the soils and rhizospheres of two species of Solanum
- PMID: 31923250
- PMCID: PMC6953851
- DOI: 10.1371/journal.pone.0227422
Diversity and plant growth-promoting functions of diazotrophic/N-scavenging bacteria isolated from the soils and rhizospheres of two species of Solanum
Abstract
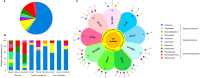
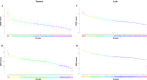
Studies of the interactions between plants and their microbiome have been conducted worldwide in the search for growth-promoting representative strains for use as biological inputs for agriculture, aiming to achieve more sustainable agriculture practices. With a focus on the isolation of plant growth-promoting (PGP) bacteria with ability to alleviate N stress, representative strains that were found at population densities greater than 104 cells g-1 and that could grow in N-free semisolid media were isolated from soils under different management conditions and from the roots of tomato (Solanum lycopersicum) and lulo (Solanum quitoense) plants that were grown in those soils. A total of 101 bacterial strains were obtained, after which they were phylogenetically categorized and characterized for their basic PGP mechanisms. All strains belonged to the Proteobacteria phylum in the classes Alphaproteobacteria (61% of isolates), Betaproteobacteria (19% of isolates) and Gammaproteobacteria (20% of isolates), with distribution encompassing nine genera, with the predominant genus being Rhizobium (58.4% of isolates). Strains isolated from conventional horticulture (CH) soil composed three bacterial genera, suggesting a lower diversity for the diazotrophs/N scavenger bacterial community than that observed for soils under organic management (ORG) or secondary forest coverture (SF). Conversely, diazotrophs/N scavenger strains from tomato plants grown in CH soil comprised a higher number of bacterial genera than did strains isolated from tomato plants grown in ORG or SF soils. Furthermore, strains isolated from tomato were phylogenetically more diverse than those from lulo. BOX-PCR fingerprinting of all strains revealed a high genetic diversity for several clonal representatives (four Rhizobium species and one Pseudomonas species). Considering the potential PGP mechanisms, 49 strains (48.5% of the total) produced IAA (2.96-193.97 μg IAA mg protein-1), 72 strains (71.3%) solubilized FePO4 (0.40-56.00 mg l-1), 44 strains (43.5%) solubilized AlPO4 (0.62-17.05 mg l-1), and 44 strains produced siderophores (1.06-3.23). Further, 91 isolates (90.1% of total) showed at least one PGP trait, and 68 isolates (67.3%) showed multiple PGP traits. Greenhouse trials using the bacterial collection to inoculate tomato or lulo plants revealed increases in plant biomass (roots, shoots or both plant tissues) elicited by 65 strains (54.5% of the bacterial collection), of which 36 were obtained from the tomato rhizosphere, 15 were obtained from the lulo rhizosphere, and 14 originated from samples of soil that lacked plants. In addition, 18 strains showed positive inoculation effects on both Solanum species, of which 12 were classified as Rhizobium spp. by partial 16S rRNA gene sequencing. Overall, the strategy adopted allowed us to identify the variability in the composition of culturable diazotroph/N-scavenger representatives from soils under different management conditions by using two Solanum species as trap plants. The present results suggest the ability of tomato and lulo plants to enrich their belowground microbiomes with rhizobia representatives and the potential of selected rhizobial strains to promote the growth of Solanum crops under limiting N supply.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

