VGLUT substrates and inhibitors: A computational viewpoint
- PMID: 31923412
- PMCID: PMC7338262
- DOI: 10.1016/j.bbamem.2020.183175
VGLUT substrates and inhibitors: A computational viewpoint
Abstract
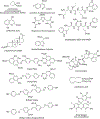
The vesicular glutamate transporters (VGLUTs) bind and move glutamate (Glu) from the cytosol into the lumen of synaptic vesicles using a H+-electrochemical gradient (ΔpH and Δψ) generated by the vesicular H+-ATPase. VGLUTs show very low Glu binding and to date, no three-dimensional structure has been elucidated. Prior studies have attempted to identify the key residues involved in binding VGLUT substrates and inhibitors using homology models and docking experiments. Recently, the inward and outward oriented crystal structures of d-galactonate transporter (DgoT) emerged as possible structure templates for VGLUT. In this review, a new homology model for VGLUT2 based on DgoT has been developed and used to conduct docking experiments to identify and differentiate residues and binding orientations involved in ligand interactions. This review describes small molecule-ligand interactions including docking using a VGLUT2 homology model derived from DgoT.
Keywords: Docking; Glutamate; Homology models; Inhibitor; Substrate; Vesicular glutamate transporter.
Copyright © 2020. Published by Elsevier B.V.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





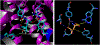
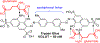
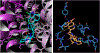
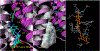
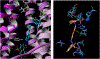

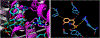


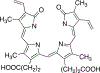

References
-
- Zhang FX, Ge SN, Dong YL, Shi J, Feng YP, Li Y, Li YQ, Li JL, Vesicular glutamate transporter isoforms: the essential players in the somatosensory systems, Prog. Neurobiol 171 (2018) 72–89. - PubMed
-
- Omote H, Miyaji T, Juge N, Moriyama Y, Vesicular neurotransmitter transporter: bioenergetics and regulation of glutamate transport, Biochemistry 50 (2011) 5558–5565. - PubMed
-
- El Mestikawy S, Wallen-Mackenzie A, Fortin GM, Descarries L, Trudeau LE, From glutamate co-release to vesicular synergy: vesicular glutamate transporters, Nat. Rev. Neurosci 12 (2011) 204–216. - PubMed
-
- Liguz-Lecznar M, Skangiel-Kramska J, Vesicular glutamate transporters (VGLUTs): the three musketeers of glutamatergic system, Acta Neurobiol. Exp. (Wars) 67 (2007) 207–218. - PubMed
-
- Takamori S, VGLUTs: ‘exciting’ times for glutamatergic research? Neurosci. Res 55 (2006) 343–351. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

