An unsupervised learning approach to identify novel signatures of health and disease from multimodal data
- PMID: 31924279
- PMCID: PMC6953286
- DOI: 10.1186/s13073-019-0705-z
An unsupervised learning approach to identify novel signatures of health and disease from multimodal data
Abstract
Background: Modern medicine is rapidly moving towards a data-driven paradigm based on comprehensive multimodal health assessments. Integrated analysis of data from different modalities has the potential of uncovering novel biomarkers and disease signatures.
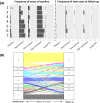
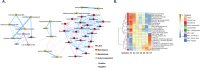
Methods: We collected 1385 data features from diverse modalities, including metabolome, microbiome, genetics, and advanced imaging, from 1253 individuals and from a longitudinal validation cohort of 1083 individuals. We utilized a combination of unsupervised machine learning methods to identify multimodal biomarker signatures of health and disease risk.
Results: Our method identified a set of cardiometabolic biomarkers that goes beyond standard clinical biomarkers. Stratification of individuals based on the signatures of these biomarkers identified distinct subsets of individuals with similar health statuses. Subset membership was a better predictor for diabetes than established clinical biomarkers such as glucose, insulin resistance, and body mass index. The novel biomarkers in the diabetes signature included 1-stearoyl-2-dihomo-linolenoyl-GPC and 1-(1-enyl-palmitoyl)-2-oleoyl-GPC. Another metabolite, cinnamoylglycine, was identified as a potential biomarker for both gut microbiome health and lean mass percentage. We identified potential early signatures for hypertension and a poor metabolic health outcome. Additionally, we found novel associations between a uremic toxin, p-cresol sulfate, and the abundance of the microbiome genera Intestinimonas and an unclassified genus in the Erysipelotrichaceae family.
Conclusions: Our methodology and results demonstrate the potential of multimodal data integration, from the identification of novel biomarker signatures to a data-driven stratification of individuals into disease subtypes and stages-an essential step towards personalized, preventative health risk assessment.
Keywords: Cardiometabolic syndrome; Metabolomics; Multimodal; Network analysis; Preventative medicine; Unsupervised machine learning.
Conflict of interest statement
IS, ETC, LH, LAN, RRH, MH, IVC, HCY, CLS, NMS, WL, KEN, PB, AMK, CTC, JCV, DSK, EFK, and NS are past or current employees or contractors of Human Longevity, Inc. The remaining authors declare that they have no competing interests.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

