The RNA-binding protein HuR is a negative regulator in adipogenesis
- PMID: 31924774
- PMCID: PMC6954112
- DOI: 10.1038/s41467-019-14001-8
The RNA-binding protein HuR is a negative regulator in adipogenesis
Abstract
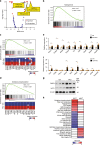
Human antigen R (HuR) is an essential regulator of RNA metabolism, but its function in metabolism remains unclear. This study identifies HuR as a major repressor during adipogenesis. Knockdown and overexpression of HuR in primary adipocyte culture enhances and inhibits adipogenesis in vitro, respectively. Fat-specific knockout of HuR significantly enhances adipogenic gene program in adipose tissues, accompanied by a systemic glucose intolerance and insulin resistance. HuR knockout also results in depot-specific phenotypes: it can repress myogenesis program in brown fat, enhance inflammation program in epidydimal white fat and induce browning program in inguinal white fat. Mechanistically, HuR may inhibit adipogenesis by recognizing and modulating the stability of hundreds of adipocyte transcripts including Insig1, a negative regulator during adipogenesis. Taken together, our work establishes HuR as an important posttranscriptional regulator of adipogenesis and provides insights into how RNA processing contributes to adipocyte development.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

