Dynamic Competition of Polycomb and Trithorax in Transcriptional Programming
- PMID: 31928411
- PMCID: PMC7311296
- DOI: 10.1146/annurev-biochem-120219-103641
Dynamic Competition of Polycomb and Trithorax in Transcriptional Programming
Abstract
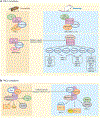
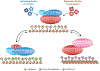
Predicting regulatory potential from primary DNA sequences or transcription factor binding patterns is not possible. However, the annotation of the genome by chromatin proteins, histone modifications, and differential compaction is largely sufficient to reveal the locations of genes and their differential activity states. The Polycomb Group (PcG) and Trithorax Group (TrxG) proteins are the central players in this cell type-specific chromatin organization. PcG function was originally viewed as being solely repressive and irreversible, as observed at the homeotic loci in flies and mammals. However, it is now clear that modular and reversible PcG function is essential at most developmental genes. Focusing mainly on recent advances, we review evidence for how PcG and TrxG patterns change dynamically during cell type transitions. The ability to implement cell type-specific transcriptional programming with exquisite fidelity is essential for normal development.
Keywords: Polycomb; Trithorax; bivalent chromatin; epigenetics; gene regulation.
Figures



Similar articles
-
DNA sequence models of genome-wide Drosophila melanogaster Polycomb binding sites improve generalization to independent Polycomb Response Elements.Nucleic Acids Res. 2019 Sep 5;47(15):7781-7797. doi: 10.1093/nar/gkz617. Nucleic Acids Res. 2019. PMID: 31340029 Free PMC article.
-
From genetics to epigenetics: the tale of Polycomb group and trithorax group genes.Chromosome Res. 2006;14(4):363-75. doi: 10.1007/s10577-006-1069-y. Chromosome Res. 2006. PMID: 16821133 Review.
-
Functional conservation of Asxl2, a murine homolog for the Drosophila enhancer of trithorax and polycomb group gene Asx.PLoS One. 2009;4(3):e4750. doi: 10.1371/journal.pone.0004750. Epub 2009 Mar 9. PLoS One. 2009. PMID: 19270745 Free PMC article.
-
Polycomb and Trithorax Group Genes in Drosophila.Genetics. 2017 Aug;206(4):1699-1725. doi: 10.1534/genetics.115.185116. Genetics. 2017. PMID: 28778878 Free PMC article. Review.
-
Histone trimethylation and the maintenance of transcriptional ON and OFF states by trxG and PcG proteins.Genes Dev. 2006 Aug 1;20(15):2041-54. doi: 10.1101/gad.388706. Genes Dev. 2006. PMID: 16882982 Free PMC article.
Cited by
-
Normal Patterns of Histone H3K27 Methylation Require the Histone Variant H2A.Z in Neurospora crassa.Genetics. 2020 Sep;216(1):51-66. doi: 10.1534/genetics.120.303442. Epub 2020 Jul 10. Genetics. 2020. PMID: 32651262 Free PMC article.
-
Clinical Correlations of Polycomb Repressive Complex 2 in Different Tumor Types.Cancers (Basel). 2021 Jun 24;13(13):3155. doi: 10.3390/cancers13133155. Cancers (Basel). 2021. PMID: 34202528 Free PMC article.
-
Rbbp4 Suppresses Premature Differentiation of Embryonic Stem Cells.Stem Cell Reports. 2021 Mar 9;16(3):566-581. doi: 10.1016/j.stemcr.2021.01.009. Epub 2021 Feb 18. Stem Cell Reports. 2021. PMID: 33606987 Free PMC article.
-
Evidence for dual roles of histone H3 lysine 4 in antagonizing Polycomb group function and promoting target gene expression.Genes Dev. 2024 Nov 27;38(21-24):1033-1046. doi: 10.1101/gad.352181.124. Genes Dev. 2024. PMID: 39562140 Free PMC article.
-
The PRC2.1 subcomplex opposes G1 progression through regulation of CCND1 and CCND2.Elife. 2025 Feb 4;13:RP97577. doi: 10.7554/eLife.97577. Elife. 2025. PMID: 39903505 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

