The Escherichia coli RnlA-RnlB toxin-antitoxin complex: production, characterization and crystallization
- PMID: 31929184
- PMCID: PMC6957109
- DOI: 10.1107/S2053230X19017175
The Escherichia coli RnlA-RnlB toxin-antitoxin complex: production, characterization and crystallization
Abstract
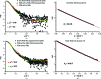
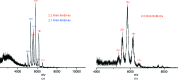
The Escherichia coli rnlAB operon encodes a toxin-antitoxin module that is involved in protection against infection by bacteriophage T4. The full-length RnlA-RnlB toxin-antitoxin complex as well as the toxin RnlA were purified to homogeneity and crystallized. When the affinity tag is placed on RnlA, RnlB is largely lost during purification and the resulting crystals exclusively comprise RnlA. A homogeneous preparation of RnlA-RnlB containing stoichiometric amounts of both proteins could only be obtained using a His tag placed C-terminal to RnlB. Native mass spectrometry and SAXS indicate a 1:1 stoichiometry for this RnlA-RnlB complex. Crystals of the RnlA-RnlB complex belonged to space group C2, with unit-cell parameters a = 243.32, b = 133.58, c = 55.64 Å, β = 95.11°, and diffracted to 2.6 Å resolution. The presence of both proteins in the crystals was confirmed and the asymmetric unit is likely to contain a heterotetramer with RnlA2:RnlB2 stoichiometry.
Keywords: Escherichia coli; bacterial stress response; macromolecular complex; toxin–antitoxin.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

